Project X
From 2006.igem.org
Back to ETH Zurich main page.
Back to the Development Group (the group that developed the project detailed below).
Contents |
Introduction
Here you find the current development stage of our project variant called "Project X", which is a result based on the initial project variants.
Complexity of behavior vs. simplicity of design
As for any project, there is obviously a trade-off: on one hand we would like to have complex behavior which makes the project interesting but also increases workload and risk of failure, on the other hand we want to choose the simplest possible and thus most elegant design in order to simplify the design task and to increase the overall feasability.
Aspect of submodules
For iGEM, there is a third consideration: to include submodules that are interesting in themselves, such as generation control, AND-dependencies, cellulose production control etc. (see details below). Each of these modules alone could prove to be an interesting contribution.
Role of this concept draft
For now, we have chosen to describe an obviously rather complex variant. We are well aware that we could greatly simplify the mechanisms and still get a similar behavior if we do choose not to implement all the modules mentioned above and detailed below.
However, it is the task of the whole team to make those evaluations and the resulting adaptations, i.e. simplifications - once we have decided whether this is the general direction to go or not.
Concept
This is the draft of our current project concept. It is obviously under constant development - at least until the team meeting on Thursday, August 11th, when it will be presented.
Goals / Output
In a nutshell, two different strains of bacteria interact with each other over several complex steps in such a way, that they will form 3D-patterns which can be observed over fluorescence. In the end, hopefully a physical structure will emerge that will persist even if the organisms die. These structures will be hollow cellulose/biofilm spheres of unknown stability. On the way, we plan to use and implement several functional genetic modules that lead to specific behaviors.
Setup
Two engineered strains of (probably) [http://en.wikipedia.org/wiki/E.coli E.coli], A and B, in a tank (x,y,z). The population of A is initially much larger than the population of B.
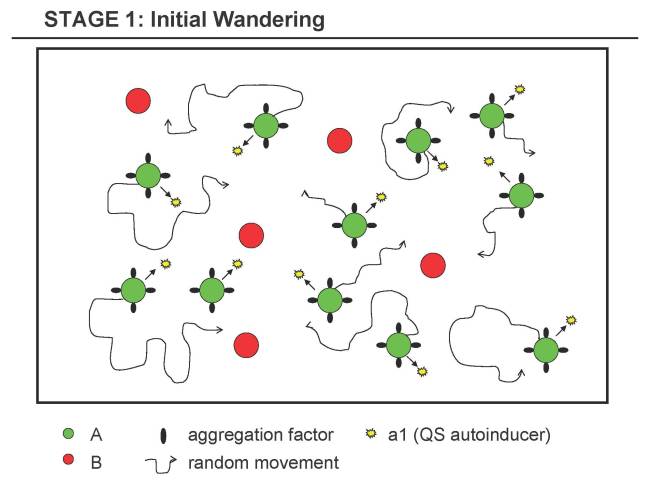
Stage 1 - Initial Wandering Stage
- 1.1 Both, A's and B's cell division is somehow blocked (or naturally very low, but can be increased in a controlled way for B).
- 1.2 A constitutively expresses aggregation factor for A, i.e. A tends to stick to A when encountered (when cells tumble into each other on their random course).
- 1.3 A constitutively expresses some signaling substance a1 for [http://en.wikipedia.org/wiki/Quorum_sensing Quorum Sensing], i.e. A can sense its own (local) density.
- 1.4 A and B constitutively express fluorescence genes (different colors) for debugging purposes (true for all steps detailed below). Alternatively, if the expression cost is too high or not suitable, specific dyes could be used (that allow to differentiate between the A and B strain).
- Comments:
- C1.1 Note: changed constitutive expression of b1 to Quorum Sensing of A (now 1.3).
- C1.2 Cell division control: If we can actually achieve cell division control, i.e. blocking at this stage, then this could be an interesting contribution. But obviously, there are simpler solutions to achieve the same goal (i.e. indirect means, see Q1.3).
- C1.3 Probably, the B population should be much smaller than the A population, so that B does not interfere with the clustering of A (i.e. not hampering the aggregation and no inclusions of B-cells within the A cluster).
- Questions:
- Q1.1 What aggregation factor to use, and how well would it work, i.e. how close do the cells have to be in order to "get into touch" and how strong will the attachment be (e.g. in the case when the contents of the tank are stirred).
- Q1.2 If the aggregation rate is low, would chemotaxis help (probably even harder to do)? What about stirring the contents of the tank? At what speed if any?
- Q1.3 How can the cell division be inhibited in a direct way (as opposed to using indirect means, such as restriction of substrate, poisoning, lack of enzymes)? Is it feasable (for us within the context of iGEM)?
- Q1.4 What mechanism/part to use for quorum sensing? Are there suitable standard parts in the library?
- Answers:
- A1.1: There are different candidates for bacterial aggregation: Ag43, Type1 Fimbriae and TibA. They are surface structures that can self-recognize and trigger aggregation and flocculation. Most of the aggregation tests for these factors have been done on plates or in static liquid culture. It is still unclear whether they would be active for stirred solution but there are certainly parameters such as the speed and the temperature to play around in order to improve the aggregation. The time that passes before aggregation is initiated is highly dependent on the cell density of the initial suspension. It seems to be in good agreement with a model following first-order kinetics, where the chance of two bacteria colliding at a given time interval is proportional to the cell density (Hasman et al. 1999). According to the publication of Sherlock et al. (2005), it seems that TibA Adhesin/Invasin would also trigger aggregation in a static culture and then settling of the bacterial culture to the bottom part. One can imagine to start with static culture and to shift slowly to non-static culture to speed up the aggregation process. Working with static culture can also be easier to predict. Have a look on the static mixed culture of bacteria expressing and non-expressing AIDA adhesin from Sherlock publication fig.5B (Sherlock et al. 2004).
- A1.2: I personally think that would be more difficult to achieve. You would have to get just some cells to express an attractant, toward which the others will move. I think that is not easy to implement (or even impossible?) in a stirred, liquid medium.
- A1.4: A very well studied quorum sensing system is that of V. fisheri, that results in bioluminescence. The genes for making luciferase, and therefore bioluminescence, are contained in the lux operon. LuxR, the transcriptional activator, is constitutively transcribed at a low level and binds to the lux operon (luxO) near the lux promoter (luxP). LuxI, an HSL (homoserine-lactone), is the inducer. It is produced at basal level by every cell and can diffuse out of the cell membrane. At low cell density LuxI readily diffuse out of the cells, whereas at high cell density it accumulates at an intracellular concentration which equals the extracellular one. At such high local concentrations, LuxI binds to LuxR, enabling it to bind to the operator and turn on transcription of the lux operon. The parts for this quorum sensing system are already in the registry ([http://parts2.mit.edu/r/parts/partsdb/view.cgi?part_id=192],[http://parts2.mit.edu/r/parts/partsdb/view.cgi?part_id=193]). Other quorum sensing systems exist, which could be used as second or third communication module, since the dedicated HSL (autoninducers) will not interfere with non-cognate systems. At least two of these alternative systems (RhlR-RhlI and LasR-LasI from P.aeruginosa) are available as BioBricks in the registry.
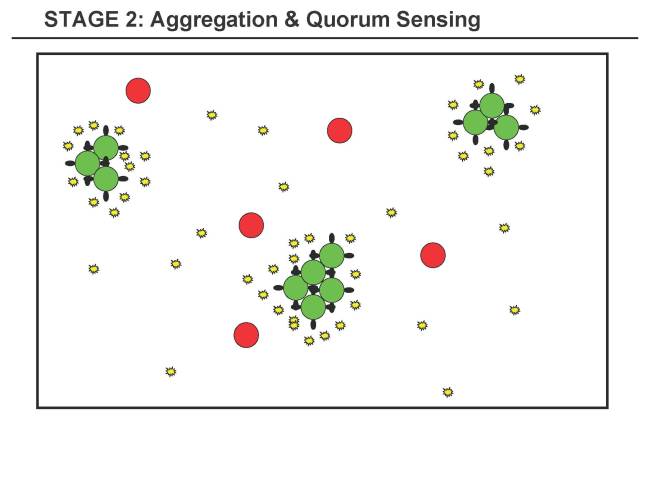
Stage 2 - Aggregation & Quorum Sensing Stage
- 2.1 A starts to aggregate and form clusters of A-cells (not necessarily spherical).
- 2.2 A senses the increasing concentration of substance a1 within the cluster
- Comments:
- C2.1 Note: changed former point "2.2 B senses concentration of substance a1" to Quorum Sensing of A
- C2.2 B is not involved at this point at all (as opposed to before). This might hopefully increase robustness, i.e. make it more likely that we can actually achieve the desired behavior.
- C2.3 Clustering would be useful in many contexts and it is something we could easily observe (debugging).
- Questions:
- Q2.1 Tuning of Quorum Sensing, i.e. when the threshold of a1 is reached in A-cells and taking into account where it is reached first (probably in the center of the A-cluster).
- Q2.2 The aggregation needs to be faster than the increase of a1 or a1 has to degrade in order to form a gradient (so that A do not start triggering B before proper clustering). How well can this be tuned? Another solution could be that the quorum sensing only starts after aggregation, e.g. that cell-to-cell contact triggers the production of a1.
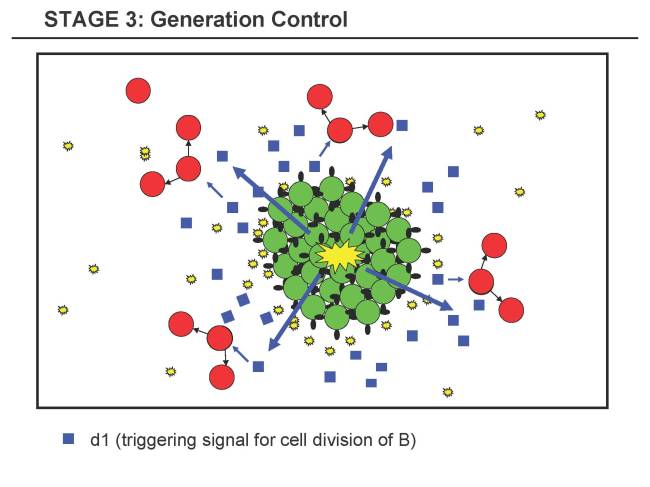
Stage 3 - Generation Control
- 3.1 The clusters of A reach a critical size. This leads to a certain concentration of a1 within the cluster (Quorum Sensing).
- 3.2 The threshold of a1 is reached and in A a division signal d1 for B is produced. This triggers intensive cell division of B in the vicinity.
- Comments:
- C3.1 The reasons for this rather complicated approach are: i) We have true Quorum Sensing of A, ii) We have the behavior of B fully decoupled from A up to this point (lower risk of B locally sensing too high concentrations of a1 and prematurely starting cell devision).
- C3.2 The clusters A will tend to have a specific size, which can be tuned over adjusting the sensitivity of A to a1. This is a cool (and hopefully observable) side-effect.
- Questions:
- Q3.1 Tuning of Quorum Sensing, i.e. defining the threshold of a1 and possibly degradation of a1.
- Q3.2 How to trigger cell division in B over some substance d1? Is it feasable? (otherwise we can use enzyme-based approaches as proposed by Herve).
- Q3.3 How well can d1 diffuse out of the A-cluster?
- Q3.4 Will there be enough B cells in the vicinity sensing d1?
- Q3.5 What shape will the clusters have at this point? Rather spherical or basically random shapes? Will this interfere with the outcome?
- Q3.6 What will be the behavior of the A- and B-clusters? Obviously they will have higher inertia. Is this an advantage or a disadvantage? How will this affect the overall behavior? Will they sink to the ground for some reason?
- Q3.7 It is important that d1 does not interfere with quorum sensing of A. Are there useful signaling molecules other than oxodecanoyl AHL?
- Answers:
- A3.2: General Information and possible alternatives:
- Replication inhibitors: By impeding DNA replication, there is a possibility to maintain the cell in a non-multiplying state. For example, SeqA competes with initiation complex (binding of dnaA to the oriC then DNA double helix separation then prepriming complex orientation by dnaA) (Torheim et al., 1999). SeqA appears also to release dnaA from the oriC (Taghbalout et al. 2000). The problem with the SeqA approach is that there is little information about the expression that is required to really obtain an inhibitory effect on the initiation of replication. Most of the studies have been done in vitro. This would then be a new challenge if we try to stop bacterial growth with this approach or at least we could probably increase the generation time but then again it is difficult to predict.
- Cell division inhibitors: The idea would be to express an inhibitor of the cell division in the population that we want to maintain small in the first phase of the process. It would certainly slow down the bacterial growth but they would keep growing so it cannot be considered then as a tight growth control. It is also difficult to predict but it forms structures (filaments) that might be of interest. Here are two examples of cell division inhibitors
- SulA = SfiA: Part of the SOS response (DNA repair system).Inhibition of cell division after DNA damage (Justice, Garcia-Lara et al. 2000) Over expression of SulA prevents the formation of ftz ring at the mid cell (Mukherjee, Cao et al. 1998) SulA appears to block the septation by interfering with FtsZ polymerization and, consequently, preventing FtsZ ring formation.
- MinCD: MinCD division inhibitor is part of the MinCDE system that determines the correct placement of the division septum. (Justice, Garcia-Lara et al. 2000) Overexpression of MinC and MinD lead to the inhibition of the cell division at all potential division sites (de Boer, Crossley et al. 1989).
- Sugar mutants: The idea is to use a mutant population B that cannot grow in the medium. The mutant bacteria population lacks an enzyme to process the carbohydrate source. This enzyme would be under an inducible promoter that could be trigger whn the population A is over with its job.
- PTS: MutantsP-enolpyruvate (in) + carbohydrate (out) ---PTS---> pyruvate (in) + carbohydrate-P (in) Carbohydrate phosphorylation is coupled to its translocation across the membrane, the energy for these processes being provided by the glycolytic intermediate PEP (Postma93). In bacteria, a large number of sugars and hexitols are transported by the phosphorenolpyruvate (PEP)-dependent phosphotransferase system (PTS). The ptsHI genes from Streptococcus mutans and Staphylococcus carnosu were shown to complement E.coli pts mutants (Kohlbrecher, Eisermann et al. 1992; Boyd, Cvitkovitch et al. 1994). Pts E.coli mutants don’t grow on PTS sugars such as glucose, mannose, mannitol.
- A3.7: There is a range of other AHL known, and they are known to be uncapable of cross stimulation of noncognate systems (miller01). Alternative system are available in the registry.
- A3.2: General Information and possible alternatives:
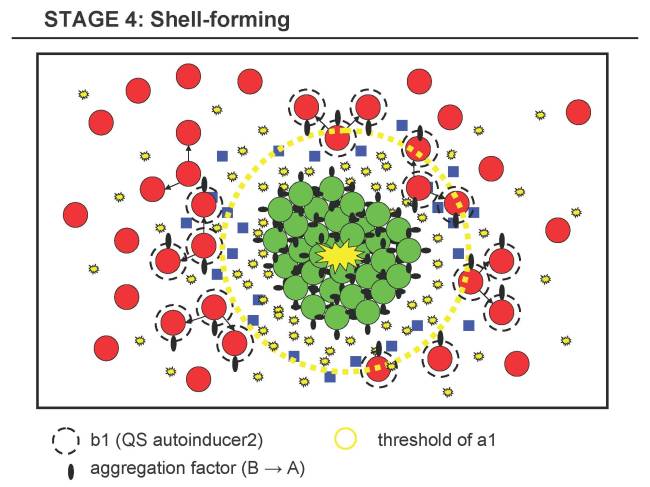
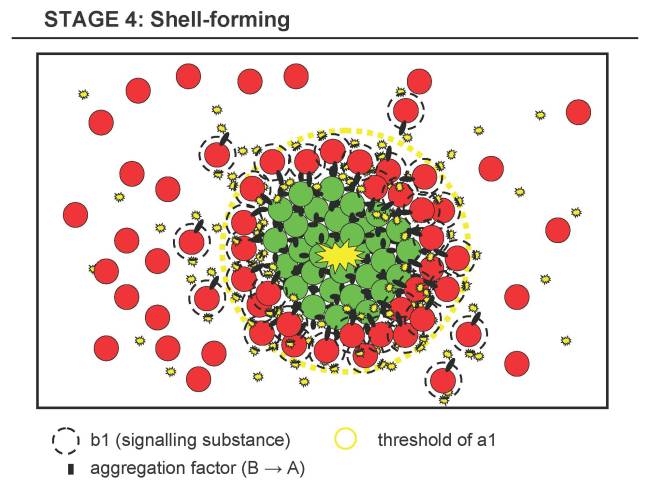
Stage 4 - Shell-forming Stage
- 4.1 B is sensitive to a certain concentration of a substance diffusing from the cluster, e.g. a1, which should have a high degradation rate (and thus forming a steep gradient).
- 4.2 Thus, only B-cells close to the A-cluster can sense the threshold of a1 and will then express the same aggregation factor that makes B attach to A or B, slowly forming a shell of B around A of increasing thickness. B-cells below this threshold will not start to aggregate and keep floating around.
- 4.3 In parallel, B possibly expresses a signalling substance b1.
- Comments:
- C4.1 The increasing thickness of the shell will probably hamper the diffusion of d1 and thus the cell division of B should stop.
- Questions:
- Q4.1 Tuning of the steepness of the gradient of a1, and thus tuning the degradation rate of a1.
- Q4.2 Probability that B-cells will rather attach to A-cluster than other B-cells.
- Q4.3 Will there be enough B cells in the vicinity sensing d1? Will the cells in the vicinity ever stop to divide, and if not, will this pose a problem?
- Q4.4 What shape will the clusters with their shells have at this point? Rather spherical or basically random shapes? Will this interfere with the outcome?
- Answers:
- A4.X: -
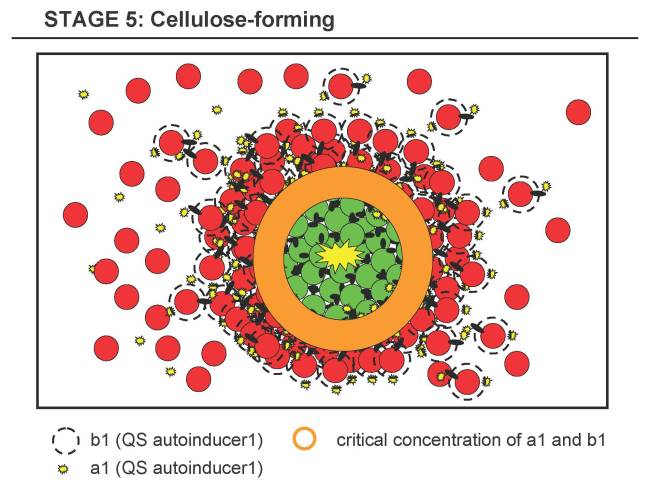
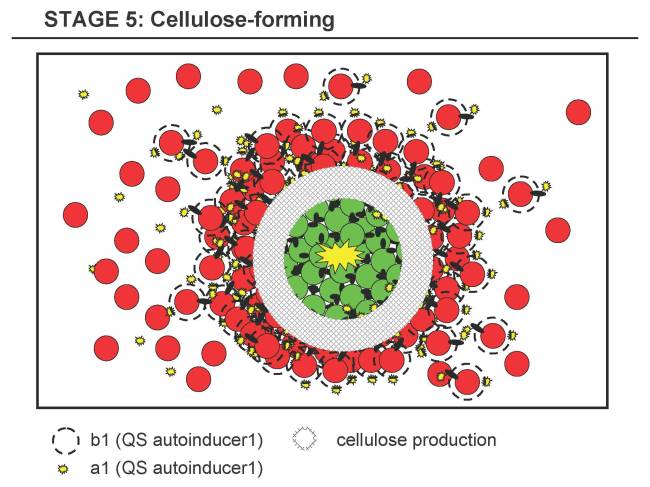
Stage 5 - Cellulose-forming Stage
- 5.1 The shell of B forms around the clusters of A and reaches a certain density at the A-B-interface (shell-core boundary).
- 5.2 a1 and b1 reach certain concentrations at the A-B-interface (or the docking of B to A might trigger some response), which in turn triggers the production of cellulose in both types, A and B. Either as AND-implementation (a1 and b1 have both to be present at certain thresholds) or simply just by b1 triggering A and a1 triggering B (while both are degrading rapidly, forming a gradient) or even simpler by skipping the necessity of a1 or b1 and/or only implementing cellulose production in one strain.
- Comments:
- C5.1 Note: changing the former steps to Quorum Sensing of A leads to decoupling etc., yes, but it has the clear disadvantage (or is it an advantage?) that now the interface-sensing based on some quickly degrading substances a1 and b1 is also decoupled, i.e. they have to be additionally produced. Thus we would gain in decoupling into independent substages and probably in robustness of the desired behavior, but we also have additional genes and cost.
- C5.2 The implementation of an AND-module is more complex, but also more sexy.
- Questions:
- Q5.1 How well can the degradation rate of a1 and b1, respectively, be tuned? Tuning is necessary, since you don't want the overlap of critical concentrations of a1 and b1 to be too wide. Ideally, this overlap would only consist of a belt of some few layers of A and B cells at the interface.
- Q5.2 How difficult is it to implement an AND-module?
- Q5.3 Can cellulose be sufficiently excreted by E.coli? Are the genes identified and could be synthesized? Would the parts excreted by different cells sufficiently attach to each other? Would the cellulose matrix be of sufficient strenght?
- Q5.4 Will cellulose be produced in liquid environment?
- Q5.5 Will the B cells form a layer that is actually thicker than one cell? Why?
- Answers:
- A5.1 Assuming that the cells will aggregate properly and thus will be rather tightly packed forming themselves a considerable barrier for diffusion, the overlap could be fairly thin.
- A5.2 A typical AND gate is e.g. the lac promoter ([http://web.mit.edu/esgbio/www/pge/lac.html]). In order to allow transcription of lacZYA, the repressor LacI has not to be bound to the operator sequence. LacI undergoes a conformational change when interacting with IPTG or lactose and "falls down" from the operator sequence. Moreover, the inducer complex CAP-cAMP trigger docking of the polymerase on the lac promoter. Therefore, the presence of lactose (or IPTG) and cAMP make transcription from lac promoter possible. However, is it still questionable if we can modify such a system to make it compatible with our system. A NOT-AND gate could be assembled from already existing parts (design will follow).
- A5.3 Several strains with a biosynthesis protein exist ([http://ecocyc.org/ K12] or [http://genome.gen-info.osaka-u.ac.jp/bacteria/o157/ O157:H7], a rather unpleasant germ). Cellulose in bacteria serves to as a means to mechanical stability, protection, and adhesion to host plants. Cellulose seems to be "a significant constituent of the extracellular matrix observed during multicellular morphotype (rdar) growth" [http://biocyc.org/ECOLI/NEW-IMAGE?type=ENZYME&object=EG12259-MONOMER]. Zogaj et al. claim that cellulose synthesis can be turned on by the sole expression of [http://biocyc.org/ECOLI/NEW-IMAGE?type=GENE&object=EG11257 adrA], leading to the formation of a highly hydrophobic network with tightly packed cells in a rigid matrix. This indicates that the structure might be fairly strong. The bcs (bacterial cellulose synthesis) operon encodes proteins essential for cellulose biosyhtnesis: [http://biocyc.org/ECOLI/NEW-IMAGE?type=GENE&object=EG12260, bcsA], [http://biocyc.org/ECOLI/NEW-IMAGE?type=ENZYME&object=EG12259-MONOMER bcsB], [http://biocyc.org/ECOLI/NEW-IMAGE?type=GENE&object=EG12258 bcsZ], and [http://biocyc.org/ECOLI/NEW-IMAGE?type=GENE&object=EG12257 bcsC] (in this order). Thus, just using some suitable strain (as opposed to synthesizing the bcs genes) in combination with the controlled expression of adrA seems to be a straightforward way to go, no? Or we could just replace the promoter responding to AdrA with a regulated one, so that we could directly switch the transcription of the cluster on/off. However, in order to insure thight bundling of cellulose around the cells, aggregative fimbreiae have to be co-expressed with cellulose. Otherwise bundles of cellulose freely protrude from cell clumps.
- A5.4 From römling02 it seems that cellulose is produced at the air-liquid boundary. Anyway, cellulose is highly hydrophilic, so that should not be a problem.
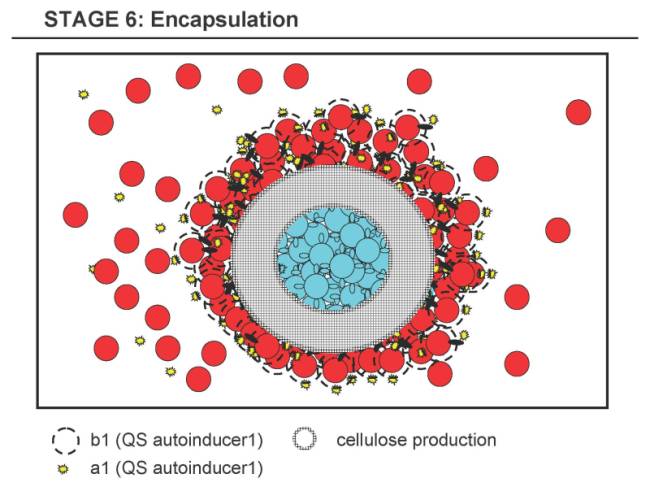
Stage 6 - Encapsulation Stage
- 6.1 Due to the growing thickness of the cellulose shell, the A-core gets isolated, e.g. limiting the diffusion of vital substances, and starts to express some different fluorescence gene (for debugging).
- Comments:
- C6.1 The formation of closed capsules of a specific size would clearly be a cool thing - and possibly even useful.
- C6.2 The subsequent fluorescence is just for debugging and to prove the point that the encapsulation could be detected and used for other purposes, e.g. the incapsulation of drugs.
- C6.3 If the A-cells are sufficiently isolated, they could also simply starve to death.
- Questions:
- Q6.1 How can the isolation be sensed by A? Should we use a different approach?
- Q6.2 How porous is such a cellulose wall and to what substances?
- Q6.3 Do we need to actively stop the process at this stage?
Methods
For now you will find the methods for each solution variant and module we have investigated so far in the corresponding stage section above, either under "Comments" or "Answers". If this project concept will be further developed, then more details will follow, of course.
Discussion
Here you find a preliminary discussion of the current project draft. The real discussion, however, will take place at the teammeeting.
Stages
The project is devided in several stages. There is a linear dependence between these stages, i.e. stage n+1 only happens when stages 1 to n worked out as planned. However, if one of these stages fails, then it is quite easy to think of manually adjusting for it, e.g. add certain substances such as aggregation factors by hand.
- Pros and Cons
- + The stages allow for proper debugging on mechanisms over fluorescence and possibly other means.
- + Stages that fail can probably be replaced by manual adjustments, so that the project can still continue if certain modules fail.
- + Additional stages can easily be added (this is illustrated by the not really necessary stage 6).
- + The stages correspond largely to certain modules and can thus easily be subdivided into tasks for different development groups.
Modularity
The project includes several interesting modules. By modules we mean certain features (likely corresponding to some isolated plasmid) that implement the more complex and thus more interesting aspects of this project. Such modules would be Cell division control, AND-dependence, and Cellulose production, to name the obvious ones. As mentioned in the introduction, we have chosen to include as many modules as possible without too much concern about the inherent risk. They can always be dropped and replaced with simpler mechanisms to achieve the same behavior.
- Pros and Cons
- + Modules are more complex, thus more interesting and possibly more useful (e.g. could be added to the parts library)
- - Modules are more complex, harder to implement and thus risky
Feasability
We spent an enourmous amount of time to have our little meetings, discussions, and literature research. However, we are by now quite confident that at least in principle all modules mentioned above can be found, adapted or designed. We put more detailed information on this in the corresponding section of each stage. Up to now, we did not stumble over a real obstacle (apart from the obvious ones: overall complexity and limited resources). But then again, we can easily sacrifice the more complex modules with less interesting mechanisms in order to make the design simpler and to lower the risk.
- Pros and Cons
- + We found possible solutions for all modules and no major problem:
- quorum sensing: Three different quorum-sensing systems (LuxR-LuxI, LasR-LasI, RhlR-RhlI)are already available in the registry. The first system is the "classical" one, that was already implemented in "real" studies (basu05, you04) as well as by iGEM teams. The degradation rate of inducers can also be regulated by getting the cells to produce degradation enzymes.
- aggregation: Aggregation factors (e.g. Ag43, Type1 Fimbriae and TibA) are surface structures that can self-recognize and trigger aggregation and formation of clumps of cells. Many studies report successful aggregation of bacteria expressing these factors (Sherlock et al. 2004),(Hasman et al. 1999). However, these test have been done on plates or in static liquid culture. Anyhow, one can imagine to start with static culture and to shift slowly to non-static culture to speed up the aggregation process.
- cell division: Cell division could be triggered directly by expressing some proteins needed by the bacteria in order to divide. Otherwise, an indirect and simple solution would be to use PTS mutants. These are bacteria which lack the enzyme needed for sugar transport, and can therefore not grow on certain sugars (e.g. glucose, mannose,...). Growth of PTS mutants can be restored by activating expression of this enzyme by means of some signal (HSL?).
- AND: This would be a tricky part (maybe even the trickiest), but is certainly possible. Randy Rettberg kindly suggested a concept that leads to realization of AND gate composed of Quad Part Inverters (which are already available in the registry). This step still requires some hard work, which will be done if this project will be chosen.
- cellulose production: Bacterial cellulose is naturally produced by at least two sequenced E.coli strains. Genes for cellulose production are organized as an operon, and their expression is regulated by a positive regulator. Thus, by controlling the expression of this positive regulator we could switch on/off transcription of cellulose genes. Cellulose seems to be the substance of choice for producing bio-3-D structures, because it is excreted from the cells as strong microfibrils that interact whith each other by forming hydrogen bonds, thus forming extended stuctures. These are visible as a mat on top of the bacteria.
- - The complexity of the current concept with all its modules is high, thus we are likely to face many smaller and larger problems during implementation, a prominent one being the tuning at the interfaces.
- + We found possible solutions for all modules and no major problem:
Usefulness
It is rather hard to argue about the usefulness of such academic and short termed projects (summer competition). However, we think there are several ways that make this project of general interest:
- Pros and Cons
- + New modules are likely to be developed that could find their way into the parts library.
- + Generation control and cell division control is obviously an interesting direction.
- + At this point, we are not sure whether a proper AND-module actually exists (it seems that we would have to make it from other logical modules). Thus this could be an interesting contribution too.
- + Cellulose seems to be a material of great interest (well, we found out on the way...). Thus controlling cellulose production and patterning in such a way could be of general interest.
- + Some interesting self-organisation behavior could be implemented, that could be further developed to make structures grow in a desired way.
- - So far we do not know of an immediate application for some probably spongy, random cellulose spheres or sacks, filled with cells and cell debris. But the absence of proof is not proof of absence :-)
- + New modules are likely to be developed that could find their way into the parts library.
Coolness
Well, this is something to debate. But so far the feedback was great: complex self-organisation behavior leading to physical evidence ... well, don't know about you, but jeeeeez!! ... :-)
Back to the Development Group (the group that developed the project detailed above).
Back to ETH Zurich main page.