Read Through Transcription Problem
From 2006.igem.org
Read-through from the carrier vector backbone leads to uncontrolled Tet expression
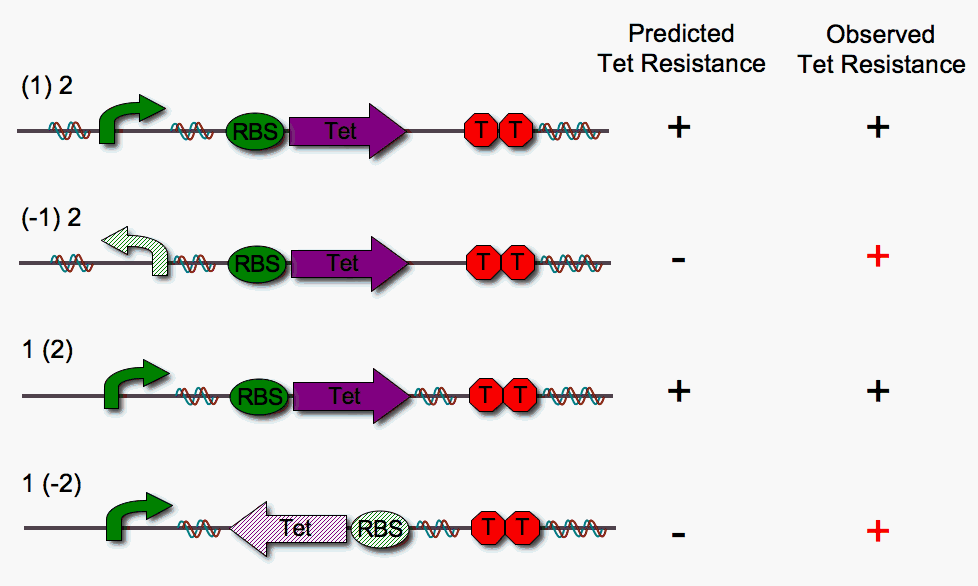
We built four different DNA pancake stacks (shown below) to test whether tetracylcine resistance is dependent upon the proper orientation of each pancake. We tested "single pancake constructs" where only one of the two units is flippable (either pBad or RBS-Tet).
As illustrated above, we anticipated that the stacks in which one pancake is in the worng orientation would fail to show tetracycline resistance. However, we observe that this is not the case. We suspect that RBS-Tet is being transcribed via a promoter within the cloning vector backbone. Read-through is a major problem, since our system requires that transcription depends strictly upon proper order and orientation of the DNA pancakes.
Read-through also leads to uncontrolled flipping
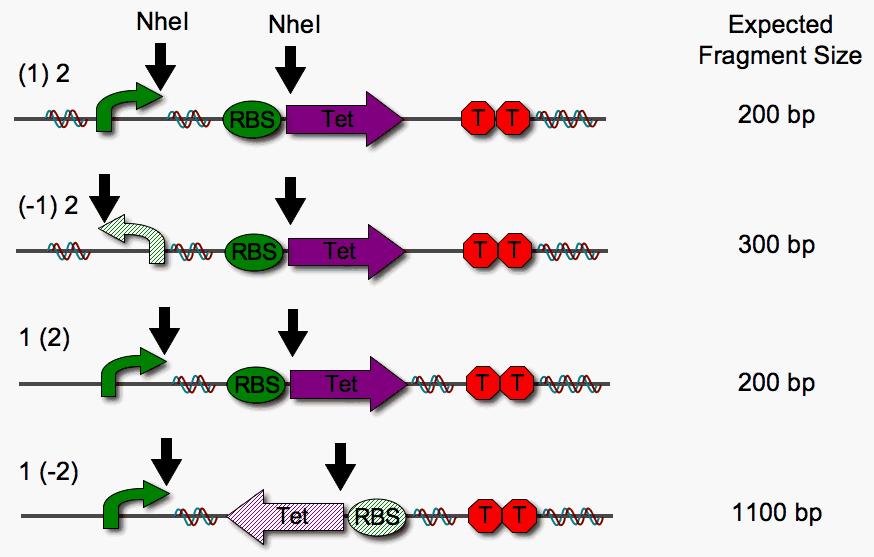
In a concurrent test to examine functionality of the Hin invertase in our system, we checked for single pancake flipping in the presence of inducible Hin expression. Since we could not use tetracycline resistance to check for orientation, we used an NheI restriction digest instead. We expected Hin expression, and therefore DNA flipping, to be dependent upon pLac induction by IPTG. However, we observe flipping without induction.
| 
|
We are excited to find that our BioBrick version of Hin is functional, but lack of control poses a problem for further studies of the kinetics of flipping. We suspect that read-through is the culprit here as well. Therefore, we designed a cloning vector to insulate our device from read-through transcription.