Rice University 2006
From 2006.igem.org
m |
m (→'''PROJECT PROPOSAL''') |
||
| (31 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
[[Image:Rice-iGEM-Logo.jpg|center]] | [[Image:Rice-iGEM-Logo.jpg|center]] | ||
| - | + | [[Image:Taxi.jpg|thumb|400px|center]] | |
<h3>Welcome</h3> | <h3>Welcome</h3> | ||
Welcome to the Rice [http://icampus.mit.edu/projects/iGem.shtml International Genetically Engineered Machine (iGEM)] Team Wiki. We are participating in the iGEM Competition for the first time so we are in the process of building our team. Currently, we are developing a 'quorumtaxis' system, which integrates the quorum-sensing and chemotaxis systems. Bacteria engineered with this quorumtaxis system will be able to move towards high concentrations of quorum pheromone. The Rice iGEM Team is divided into three groups: Experimental, Theory, and Modeling. Explore our website or [[Rice:Contact | contact us]], as we will be constantly updating with the latest developments in the project.<BR> | Welcome to the Rice [http://icampus.mit.edu/projects/iGem.shtml International Genetically Engineered Machine (iGEM)] Team Wiki. We are participating in the iGEM Competition for the first time so we are in the process of building our team. Currently, we are developing a 'quorumtaxis' system, which integrates the quorum-sensing and chemotaxis systems. Bacteria engineered with this quorumtaxis system will be able to move towards high concentrations of quorum pheromone. The Rice iGEM Team is divided into three groups: Experimental, Theory, and Modeling. Explore our website or [[Rice:Contact | contact us]], as we will be constantly updating with the latest developments in the project.<BR> | ||
| Line 31: | Line 31: | ||
[[user:Kcox|Ken Cox]] - Chemical & Biomolecular Engineering <br> | [[user:Kcox|Ken Cox]] - Chemical & Biomolecular Engineering <br> | ||
[[user:Scox|Steve Cox]] - Computational & Applied Mathematics <br><br> | [[user:Scox|Steve Cox]] - Computational & Applied Mathematics <br><br> | ||
| + | |||
| + | <br> | ||
| + | =='''PROJECT PROPOSAL'''== | ||
| + | '''Quorumtaxis''' -- The integration of quorum-sensing and chemotaxis pathways, to produce an ''E. coli'' strain which is capable of detection and movement towards concentrations of the ''B. subtilis'' quorum phermone, ComX. Upon successful quorumtaxis towards the target, the ''E. coli'' will produce a signaling response (e.g., GFP production) or a kill response (e.g., the expression of gram-positive lethal proteins). A detailed proposal outline is provided in the link below. | ||
| + | |||
| + | [[Image:QT2.gif|frame|center|Seek-and-Destroy E. coli]] | ||
| + | |||
| + | *[[PROJECT PROPOSAL]] <br> | ||
| + | |||
| + | *[[2006 Project Poster]] <br> | ||
=='''EXPERIMENTAL GROUP'''== | =='''EXPERIMENTAL GROUP'''== | ||
| Line 36: | Line 46: | ||
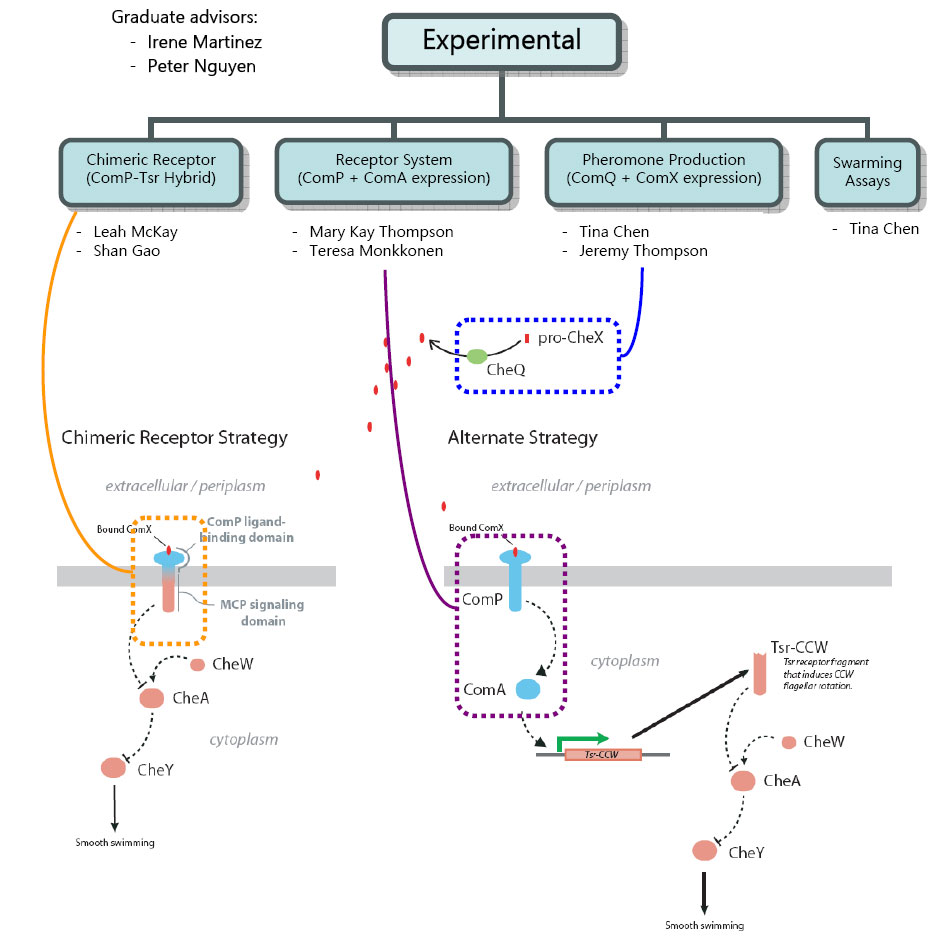
Welcome! The Rice iGEM E-group is tasked with the construction of a wholly novel phenotype -- 'quorumtaxis'. More about the specifics of this project can be found in the 'Project Proposals' section of the Rice iGEM page. Currently, we have 6 undergraduate students working on this project, with two graduate students as advisors. | Welcome! The Rice iGEM E-group is tasked with the construction of a wholly novel phenotype -- 'quorumtaxis'. More about the specifics of this project can be found in the 'Project Proposals' section of the Rice iGEM page. Currently, we have 6 undergraduate students working on this project, with two graduate students as advisors. | ||
| - | [[Image:Egroup.jpg]] | + | [[Image:Egroup.jpg|frame|center|Project Breakdown]] |
| + | =='''Experimental Work'''== | ||
| + | *[[Phase 1: PCR Amplification of Genes]] <br> | ||
| + | *[[Phase 2: Plasmid Construction and Bacterial Transformations]] | ||
| + | <br> | ||
| + | <br> | ||
| - | + | =='''MODELING GROUP'''== | |
| - | + | The mission of the modeling group is to help completely characterize "Quorumtaxis" for use as a biobrick. This means characterizing the transfer function, latency, etc. In our method, comX particles will act as an atractant that will effect the chemotaxtic pathway via the chimeric comP/MCP membrane protein. Therefore, in order to model this event, we will focu on modeling chemotaxis within bacteria. We have identified the following goals. | |
| + | # Model the response of a single bacterium to stimulus | ||
| + | # Model the phenotypic response of a bacterial colony | ||
| + | # Understand the effect of a comP/MCP chimeric protein on the chemotaxtic pathway | ||
| + | # Develop a theoretical framework of integrating the kill response in a single bacterium | ||
| + | # Simulate the kill behavior for a bacterial colony | ||
| + | ---- | ||
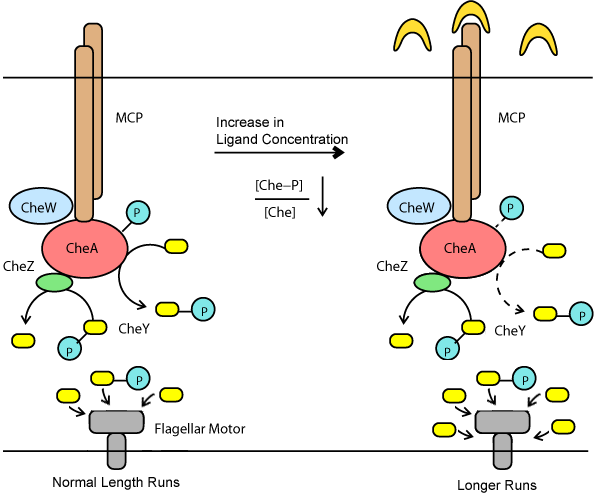
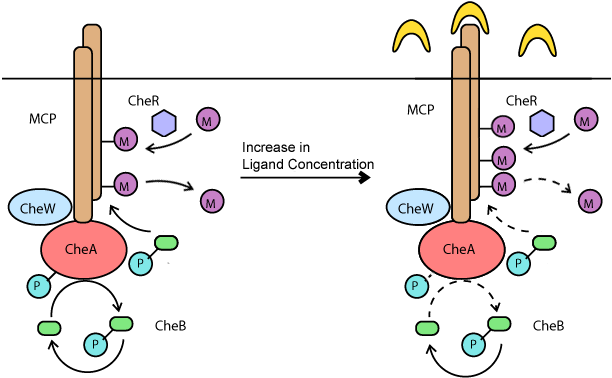
| + | Chemotaxis in bacteria is mediated by periods of long runs with tumbling. When ''E. coli'' senses a stimulant, it will change the ratio of tumbling and long-runs in response. This behavior is mediated through the chemotaxis pathway. Meythl-accepting Chemotaxis Proteins (MCP) are membrane bound proteins which bind to ligands in the periplasm. This event sets off a chain reaction of phosphorylation and meythlation reactions which result in a change of the CheY-P to CheY concentration. In turn, the ratio of CheY-P and CheY control the long-run to tumble response of a bacteria. The phosphorylation pathway mediates a quick response to stimulus that results in a long-run being favored. However, a set of slower meythlation reactions then mediate an adaptive response to desensitize the bacteria in the absense of an increasing stimulus gradient. The two figures below expalain this idea in greater detail. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | ==''' | + | [[Image:Chemo-Phospho copy.png|frame|center|A phospho-relay mediates the fast response to a stimulus.]] |
| - | + | ||
| + | |||
| + | [[Image:Chemo-Meythl copy.png|frame|center| A methyl-relay mediates a slower adapation response to a stimulus.]] | ||
| + | |||
| + | |||
| + | In order to model this system, we have used Spiro and Barakai as a basis from which to build our model. Spiro's model tries to model 12 different states between the MCP-CheW-CheA-CheZ complex which mediates different kinetic repsonse on the other protein in the pathway. The following matlab code describes the differential equations. [http://www.caam.rice.edu/~raol/model2.m] | ||
| + | <BR><BR> | ||
| + | |||
| + | |||
| + | =='''Synthetic Biology Journal Club (Summer 2006)'''== | ||
| + | ===26 June=== | ||
| + | *B. Beason gave a "tour" of the Registry and iGEM Teaching Resources and talked about the team wiki. | ||
| + | *B. Mukhopadhyay described the 2005 iGEM projects from Univ. Toronto and UC Berkeley. | ||
| - | |||
===19 June=== | ===19 June=== | ||
| - | === | + | *I. Martinez and C. Peebles described the 2005 iGEM projects from Davidson and ETH Zurich. |
| + | *T. Chen and P. Nguyen described the 2005 iGEM projects from Harvard and Univ. Texas. | ||
| + | |||
| + | ===12 June=== | ||
*J. Raol and D. Prieto described the 2005 iGEM projects from Caltech and Cambridge. | *J. Raol and D. Prieto described the 2005 iGEM projects from Caltech and Cambridge. | ||
| + | |||
===5 June=== | ===5 June=== | ||
*K.-Y. San presented ''Programmable cells: Interfacing natural and engineered gene networks'' (Kobayashi, H. et al., PNAS 101: 8414-8419, 2004). | *K.-Y. San presented ''Programmable cells: Interfacing natural and engineered gene networks'' (Kobayashi, H. et al., PNAS 101: 8414-8419, 2004). | ||
| Line 63: | Line 95: | ||
[[Image:RiceLogo.jpg|center| 125px]] | [[Image:RiceLogo.jpg|center| 125px]] | ||
| - | + | [Rice University] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Latest revision as of 04:19, 3 November 2006
Contents |
Welcome
Welcome to the Rice [http://icampus.mit.edu/projects/iGem.shtml International Genetically Engineered Machine (iGEM)] Team Wiki. We are participating in the iGEM Competition for the first time so we are in the process of building our team. Currently, we are developing a 'quorumtaxis' system, which integrates the quorum-sensing and chemotaxis systems. Bacteria engineered with this quorumtaxis system will be able to move towards high concentrations of quorum pheromone. The Rice iGEM Team is divided into three groups: Experimental, Theory, and Modeling. Explore our website or contact us, as we will be constantly updating with the latest developments in the project.
Undergraduate Students
Chris Conner - Junior, Biochemistry and Computational & Applied Mathematics
Dario Prieto - Junior, Chemical & Biomolecular Engineering
Miinkay Yu - Junior, Chemical & Biomolecular Engineering
Tina Chen - Senior, Kinesiology
Thomas Segall-Shapiro - Freshman
Teresa Monkkonen - Senior, Biochemistry
Mary Kay Thompson - Senior, Biochemistry
Shan Gao - Senior, Biochemistry
Jeremy Thompson - Senior, Biochemistry
Leah McKay - Senior, Biochemistry
Graduate Students
Bibhash Mukhopadhyay - Molecular & Cell Biology, Baylor College of Medicine
Christie Peebles - Bioengineering
Irene Martinez - Bioengineering
Jay Raol - Computational & Applied Mathematics
Peter Nguyen - Biochemistry & Cell Biology
Advisors
Beth Beason - Biochemistry & Cell Biology
George Bennett - Biochemistry & Cell Biology
Jonthan Silberg - Biochemistry & Cell Biology
Ka-Yiu San - Bioengineering
Ken Cox - Chemical & Biomolecular Engineering
Steve Cox - Computational & Applied Mathematics
PROJECT PROPOSAL
Quorumtaxis -- The integration of quorum-sensing and chemotaxis pathways, to produce an E. coli strain which is capable of detection and movement towards concentrations of the B. subtilis quorum phermone, ComX. Upon successful quorumtaxis towards the target, the E. coli will produce a signaling response (e.g., GFP production) or a kill response (e.g., the expression of gram-positive lethal proteins). A detailed proposal outline is provided in the link below.
EXPERIMENTAL GROUP
Welcome! The Rice iGEM E-group is tasked with the construction of a wholly novel phenotype -- 'quorumtaxis'. More about the specifics of this project can be found in the 'Project Proposals' section of the Rice iGEM page. Currently, we have 6 undergraduate students working on this project, with two graduate students as advisors.
Experimental Work
MODELING GROUP
The mission of the modeling group is to help completely characterize "Quorumtaxis" for use as a biobrick. This means characterizing the transfer function, latency, etc. In our method, comX particles will act as an atractant that will effect the chemotaxtic pathway via the chimeric comP/MCP membrane protein. Therefore, in order to model this event, we will focu on modeling chemotaxis within bacteria. We have identified the following goals.
- Model the response of a single bacterium to stimulus
- Model the phenotypic response of a bacterial colony
- Understand the effect of a comP/MCP chimeric protein on the chemotaxtic pathway
- Develop a theoretical framework of integrating the kill response in a single bacterium
- Simulate the kill behavior for a bacterial colony
Chemotaxis in bacteria is mediated by periods of long runs with tumbling. When E. coli senses a stimulant, it will change the ratio of tumbling and long-runs in response. This behavior is mediated through the chemotaxis pathway. Meythl-accepting Chemotaxis Proteins (MCP) are membrane bound proteins which bind to ligands in the periplasm. This event sets off a chain reaction of phosphorylation and meythlation reactions which result in a change of the CheY-P to CheY concentration. In turn, the ratio of CheY-P and CheY control the long-run to tumble response of a bacteria. The phosphorylation pathway mediates a quick response to stimulus that results in a long-run being favored. However, a set of slower meythlation reactions then mediate an adaptive response to desensitize the bacteria in the absense of an increasing stimulus gradient. The two figures below expalain this idea in greater detail.
In order to model this system, we have used Spiro and Barakai as a basis from which to build our model. Spiro's model tries to model 12 different states between the MCP-CheW-CheA-CheZ complex which mediates different kinetic repsonse on the other protein in the pathway. The following matlab code describes the differential equations. [http://www.caam.rice.edu/~raol/model2.m]
Synthetic Biology Journal Club (Summer 2006)
26 June
- B. Beason gave a "tour" of the Registry and iGEM Teaching Resources and talked about the team wiki.
- B. Mukhopadhyay described the 2005 iGEM projects from Univ. Toronto and UC Berkeley.
19 June
- I. Martinez and C. Peebles described the 2005 iGEM projects from Davidson and ETH Zurich.
- T. Chen and P. Nguyen described the 2005 iGEM projects from Harvard and Univ. Texas.
12 June
- J. Raol and D. Prieto described the 2005 iGEM projects from Caltech and Cambridge.
5 June
- K.-Y. San presented Programmable cells: Interfacing natural and engineered gene networks (Kobayashi, H. et al., PNAS 101: 8414-8419, 2004).
- P. Nguyen presented Environmentally controlled invasion of cancer cells by engineered bacteria (Anderson, J. C. et al., J. Mol. Biol. 355: 619-627, 2006).
30 May
- J. Silberg presented Directed evolution of a genetic circuit (Yokobayashi, Y. et al., PNAS 99: 16587-16591, 2002) and Programmed population control by cell-cell communication and regulated killing (You, L. et al., Nature 428: 868-871, 2004).
- S. Cox presented Construction of a genetic toggle switch in Escherichia coli (Gardner, T. et al., Nature 403: 339-342, 2000).
[Rice University]