Freiburg University 2006
From 2006.igem.org
Kouznetsov (Talk | contribs) |
|||
| (128 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | <center>'''<font color='blue'>Welcome! We are Alife Mutants.</font color>'''</center> | + | <div style="color: #ffffff; background-color: #22aadd"><br> |
| + | [[image: Earth.gif|center]] <br style="clear:both;"/> | ||
| + | {| width="96%" border="1" cellspacing="0" cellpadding="5" align="center" | ||
| + | |- | ||
| + | | | ||
| + | === <center>'''<font color='blue'>Welcome! We are Alife Mutants.</font color>'''</center> === | ||
<p> | <p> | ||
| + | This term was invented by Martin Schneider on the Rule 110 Winter Workshop in 2004 [http://www.rule110.org/amhso/index.html]. We play without rules. We discover the rules that govern life, the universe and everything to exploit these rules and to create Artificial Life. Our short-time aim was the trip to Boston in November 2006 to take a prize in the iGEM. The long term goal now is to REALLY do it in 2007! | ||
| + | <br><br> | ||
| + | '''The poster presentation at Jamboree iGEM2006 - <font color='magenta'>"Barbie Nanoatelier:</font color> Open Source DNA-nanotechnology"''' [[Media:BarbieNanoAtelier.ppt|[ppt]],[[Media:BarbieNanoAtelier.pdf|pdf]] Richard’s correction] | ||
| + | <br><br> | ||
| + | '''Summary''' | ||
| + | * we started a new branch of Biobricks depository for DNA-origami parts, devices, and systems; | ||
| + | * following Paul Rothemund [http://www.dna.caltech.edu/~pwkr/] we created an universal DNA-platform for a patterning on the nanometer scale that will allow to organize chemical reactions, assembling and qubits; | ||
| + | * we proposed a correlation spectroscopy/microscopy for the investigation of DNA folding; | ||
| + | * we designed the tetrahedral nanobot NAUTILUS, which is a 3D-DNA building block for a hierarchical assembly, nanoswarm, and amorphous computing; | ||
| + | * we suggested DNA-origami cryptography; | ||
| + | * we emphasized aesthetic principles of AL-design; and | ||
| + | * we founded "Barbie Nanoatelier: Open Source DNA-nanotechnology" that’s the way to AL and AI! | ||
| + | <br> | ||
| + | Kuznetsov A. Barbie nanoatelier // IET Synth. Biol., Vol. 1, No. 1–2, 2007, pp. 7–12. | ||
| + | [http://ieeexplore.ieee.org/iel5/4290605/4290606/04290609.pdf] | ||
| + | <br><br> | ||
| + | [[image:uf101.jpg|left|thumb|260px|The hive #101]] | ||
| + | [[image:Freiburg Team 06.jpg|left|thumb|400px|The Freiburg Team (well, at least half of it...) from left to right: Irina Petrova, Andrei Kouznetsov, Daniel Hautzinger.]] | ||
| + | |- | ||
| + | |} | ||
| - | <center> | + | === <center>The team (all stars)</center> === |
| - | + | {| width="96%" border="1" cellspacing="0" cellpadding="5" align="center" | |
| - | + | !width="32%" style="background:#7BB5EF"|Students | |
| - | + | !width="32%" style="background:#7DD5EF"|iGEM instructors | |
| - | + | !width="32%" style="background:#7FF5EF"|Faculty/staff | |
| - | + | |- | |
| - | + | | style="background:#7BB5CC" valign="top" | | |
| - | + | *[[User:Origami_Man|Origami Man]] | |
| - | == | + | |
| - | + | ||
| - | ==== | + | |
*[[User:Daniel_Hautzinger|Daniel Hautzinger]] | *[[User:Daniel_Hautzinger|Daniel Hautzinger]] | ||
*[[User:Olga_Soboleva|Olga Soboleva]] | *[[User:Olga_Soboleva|Olga Soboleva]] | ||
*[[User:Irina_Petrova|Irina Petrova]] | *[[User:Irina_Petrova|Irina Petrova]] | ||
| - | *[[User:Tim_Barrel| | + | *[[User:Tim_Barrel|Yutthaphong Phongbunchoo]] |
| - | *[[User: | + | *[[User:Monak|Mona Klein]] |
| - | + | | style="background:#7DD5CC" valign="top" | | |
| - | + | ||
*[[User:Andrei_Kouznetsov|Andrei Kouznetsov]] | *[[User:Andrei_Kouznetsov|Andrei Kouznetsov]] | ||
| - | + | *[[User:Tamara|Tamara Ulrich]] (ambassador, ETH Zurich) | |
| - | == | + | | style="background:#7FF5CC" valign="top" | |
*[[User:Svetlana_Santer|Svetlana Santer]] | *[[User:Svetlana_Santer|Svetlana Santer]] | ||
| + | *[[User:Jank|Jan Korvink]] | ||
| + | |- | ||
| + | |} | ||
| - | == The | + | === <center>The motto</center> === |
| + | {| width="96%" border="1" cellspacing="0" cellpadding="5" align="center" style="background:black" | ||
| + | |- | ||
| + | | | ||
| + | <center><span style='font-size:24.0pt;font-family:Arial;color:yellow'>Can | ||
| + | way over DNA</span></center> | ||
| + | |- | ||
| + | |} | ||
| - | === | + | === <center>The project: DNA Folding</center> === |
| + | {| width="96%" border="1" cellspacing="0" cellpadding="5" align="center" | ||
| + | |- | ||
| + | | | ||
| + | ==== No.1 - The Pipe ==== | ||
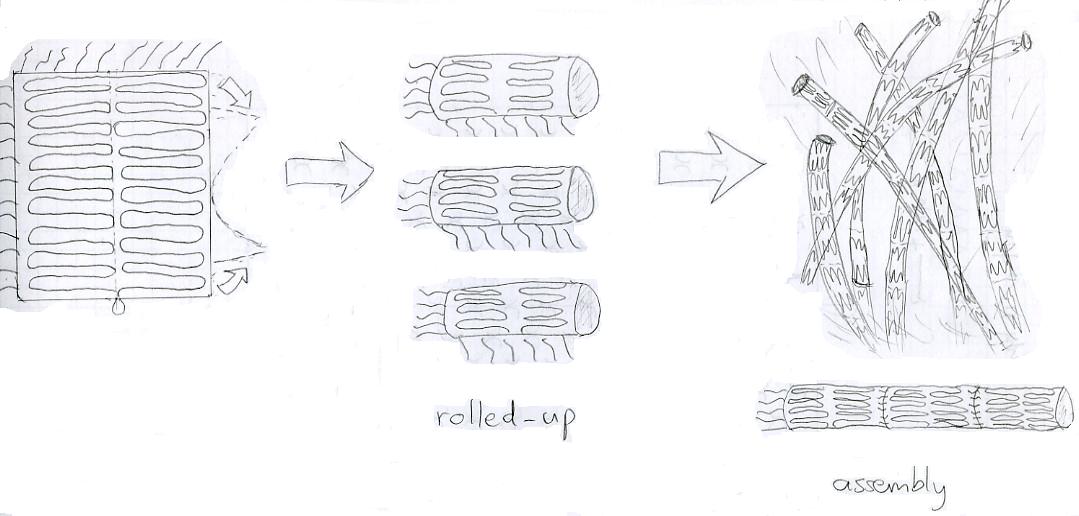
| - | = | + | [[image:123.JPG|thumb|400px|PipeLife]] |
| + | The idea is to design a strand of DNA such that it wraps into a specific shape. | ||
| + | ---- | ||
| + | There are three different stages for this project: First, the DNA should fold into a two-dimensional rectangular sheet. Secondly, this sheet should wrap itself up into the shape of a short pipe. And last, these little pipes should hook themselves up to each other such that they form one single long pipe ([[User:Tim Barrel|more details]]). | ||
| + | <br style="clear:both;"/> | ||
| + | The DNA sequence for the Pipes design can be found [http://omnibus.uni-freiburg.de/~kouznet/pipe/Tube-M13mp18-NEB-d.txt here], design picture is [http://omnibus.uni-freiburg.de/~kouznet/pipe/pip6.jpg here], the set of staples is [http://omnibus.uni-freiburg.de/~kouznet/pipe/StapleTube.txt here]. We’ll put the AFM pictures on the wiki as soon as we receive the DNA from the company... | ||
| - | + | === No.2 - Haute Couture: <font color='magenta'>Barbie Nanoatelier</font color> === | |
| - | + | [[image: iGEMBarbie.jpg|left|thumb|200px|Barbie Nanoatelier: Open Source DNA-nanotechnology]] | |
| - | + | Once the process of DNA folding into 3D structures is understood, shapes can be chosen arbitrarily. The idea of the Barbie Nanoatelier is that the DNA should wrap into a 3D T-Shirt, 3D Pants, etc. | |
| - | + | '''"sea of parts"''' | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | We need a library of molecular primitives that could be interchanged in the desirable way for a bottom-up design on the nanometer scale. Registry of DNA molecular parts and standardization is an actual question of bionanotechnology. We just put first examples of a new class of BioBricks - '''ORIGAMI BioBriks''' into [http://partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2006partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2006&group=Freiburg depository] and publish basic ideas [http://omnibus.uni-freiburg.de/~kouznet/DNA-plug.txt], [http://omnibus.uni-freiburg.de/~kouznet/T-shirt.txt], [http://omnibus.uni-freiburg.de/~kouznet/DNA-NanoSwarm.txt]. We try to [[Media:Freiburg_BB_space.pdf|merge]] a ‘dead DNA’ with an ‘alive DNA’. | |
| - | + | '''Surrealistic Science''' | |
| - | + | We also try to pump some aesthetic principles and rules (symmetry, periodic patterns, recursion, and plasticity) into our creatures. We like it because it needs a huge of imagination and really very difficult. Nobody can build a bra! - von Neumann's self-reproducing ... bra :)) | |
| - | + | ||
| - | + | ||
| - | + | ||
<br style="clear:both;"/> | <br style="clear:both;"/> | ||
| - | |||
| - | |||
| - | |||
| - | + | ==== Sketches - <font color='red'>T-Shirt from DNA</font color> ==== | |
| - | ==== | + | <gallery> |
| + | Image:TShirt1.jpg|Step 1 | ||
| + | Image:TShirt2.jpg|Step 2 | ||
| + | Image:TShirt3.JPG|Step 3 | ||
| + | Image:TShirt4.JPG|Step 4 | ||
| + | </gallery> | ||
| + | [[image:Cm-iub06.jpg|left|thumb|300px|Olga Soboleva, the presentation in Bremen sponsored by Volkswagen Stiftung]] | ||
| + | The DNA sequence for the T-Shirt design can be found [http://omnibus.uni-freiburg.de/~kouznet/t-shirt/T-shirt_M13mp18_NEB_d.txt here], the picture is [http://omnibus.uni-freiburg.de/~kouznet/t-shirt/T-shirt.jpg here], and the staples are [http://omnibus.uni-freiburg.de/~kouznet/t-shirt/T-shirt_staples.txt here] there. | ||
| + | |||
| + | '''<font color='red'>Hey, venture capitalists, where are you? The price of all of 50 billion T-Shirts would be only 7200bases x €0.17/base = €1224. Don’t miss the chance. It could be a great business tomorrow!</font color>''' | ||
| + | ---- | ||
| + | '''<font color='blue'>Ladies and Gentlemen, please, visit the demonstration of last Nike nano collection.</font color>''' [[User:Irina Petrova|(Enter)]] | ||
| + | Oh, aroma of the podium [http://omnibus.uni-freiburg.de/~kouznet/LoveParade2005.mp3 music] | ||
| + | ---- | ||
| + | '''Jobs: Still looking for catwalk models!!''' | ||
| + | <br style="clear:both;"/> | ||
| - | + | ==== No.3 - Individual projects ==== | |
| - | + | ||
| - | + | Other projects and prices could be found on individual Mutant's pages (Real iPod nano, Captain Nemo). In addition, you can find some hot information on ‘GEM Freiburg Fellow 2006’ page. The other toys will be unconventional computing, cryptography, nanoelectronics, nanooptics, drug delivery systems, smart nanomaterials, nanoswarm, eventually an artificial life and [[User:Origami_Man|Origami Man]]. Time is money! | |
| + | |- | ||
| + | |} | ||
| + | === <center>GEM Freiburg</center> === | ||
| + | {| width="96%" border="1" cellspacing="0" cellpadding="5" align="center" style="background:#CCFFFF" | ||
| + | |- | ||
| + | | | ||
| + | [[image:GEM-F-Club-tv2.jpg|right|410px]] | ||
=== Club === | === Club === | ||
| Line 75: | Line 124: | ||
=== SB Preliminary === | === SB Preliminary === | ||
| + | * [http://sommercampus2006.informatik.uni-freiburg.de/iGemAbend?action=AttachFile&do=get&target=iGemAbend.pdf iGemAbend] | ||
* DNA plug-and-play platform [http://omnibus.uni-freiburg.de/~kouznet/DNAplatform6.pdf 24.05.06] [http://omnibus.uni-freiburg.de/~kouznet/DNAplatform6-1.pdf 27.06.06] | * DNA plug-and-play platform [http://omnibus.uni-freiburg.de/~kouznet/DNAplatform6.pdf 24.05.06] [http://omnibus.uni-freiburg.de/~kouznet/DNAplatform6-1.pdf 27.06.06] | ||
* [http://omnibus.uni-freiburg.de/~kouznet/BioVLSI.pdf Very Large-Scale Integration design in Biology] | * [http://omnibus.uni-freiburg.de/~kouznet/BioVLSI.pdf Very Large-Scale Integration design in Biology] | ||
* [http://omnibus.uni-freiburg.de/~kouznet/GS-design.pdf Genetic systems design from the DNA modules] | * [http://omnibus.uni-freiburg.de/~kouznet/GS-design.pdf Genetic systems design from the DNA modules] | ||
| + | |- | ||
| + | |} | ||
| - | == Original | + | === <center>Original ideas</center> === |
| + | {| width="96%" border="1" cellspacing="0" cellpadding="5" align="center" style="background:#FFFF99" | ||
| + | |- | ||
| + | | | ||
| + | [[image:GWAL7-19.jpg|right|300px]] | ||
| + | * [[Media:Argo.pdf|Argo-machine]], [[Media:Cpm-presentation.pdf|especially for hackers]] [[Media:Argo-GWAL7-presentation.pdf|(presentation in Jena)]] | ||
| - | * [ | + | * [http://omnibus.uni-freiburg.de/~kouznet/life6000.pdf Life with a price of 6000 €] |
| - | + | Now we see it could be cheaper, but it will need a great intellectual impact. So, we’re opened for '''sponsors and relationships'''. | |
| - | === | + | === Comments === |
| - | * [http://www.edge.org/documents/archive/edge187.html#church George Church CONSTRUCTIVE BIOLOGY] - a nice paper about the challenges of modern biology | + | As I understand it, the idea is to create a "life |
| - | * [http://webcast.berkeley.edu/events/details.php?webcastid=15766 The | + | automaton" consisting of 6 proteins? That is what I took |
| + | out of your proposal. That seems quite ambitioned, but I | ||
| + | can't really judge it, and even if it is quite | ||
| + | ambitioned, that does not mean that it is bad, of | ||
| + | course. | ||
| + | |||
| + | My understanding says that you are using 6000 € for | ||
| + | denovo synthesis of 6 genes required for making minimal | ||
| + | life. Right? | ||
| + | |||
| + | So, what if after all the simulations and tests on the | ||
| + | computer, you order the DNA for minimal life and it | ||
| + | turns out that it doesn't work... What will you do then? | ||
| + | |||
| + | This is because biology is unpredictable and you must | ||
| + | not assume that the DNA which you order will work in the | ||
| + | first time itself... You must keep some backup money or | ||
| + | plans... | ||
| + | |||
| + | You have to separate specifically the general and specific | ||
| + | objectives to long and short time; implications, obstacles | ||
| + | and limitations. | ||
| + | |||
| + | OK, fine.. So you want to go in a completely different | ||
| + | direction by creating alternative life.. Good.. All the | ||
| + | best for this project. | ||
| + | |||
| + | Well, if you want to go to the extremes then are you | ||
| + | limiting yourself to already established phenomena such | ||
| + | as DNA, membranes, etc. You may want to adopt a | ||
| + | completely new and novel ideas which might be simpler | ||
| + | than DNA and other stuff of the normal cell. | ||
| + | |||
| + | So, keep all the options open. Research the literature well and create Artificial Life... | ||
| + | |||
| + | No more child labour, just molecule labour :) | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | === <center>Hey Mutant, have a look!</center> === | ||
| + | {| width="96%" border="1" cellspacing="0" cellpadding="5" align="center" style="background:#CCFFCC" | ||
| + | |- | ||
| + | | | ||
| + | === The easy and serious way. Ja oder Nein? === | ||
| + | |||
| + | * [http://www.edge.org/documents/archive/edge187.html#church George Church, CONSTRUCTIVE BIOLOGY] - a nice paper about the challenges of modern biology | ||
| + | * [http://webcast.berkeley.edu/events/details.php?webcastid=15766 The webcasts from SB2.0] [http://openwetware.org/wiki/Synthetic_Biology:Synthetic_Biology_1.0/Videos and SB1.0] | ||
| + | * [http://video.google.com/videosearch?q=iGEM iGEM related videos on Google Video] | ||
| + | |||
| + | === <font color='magenta'>DNA nanotechnology for newborn Mutants</font color> === | ||
| + | |||
| + | [http://www.npr.org/templates/story/story.php?storyId=5281562 Fun with DNA] | ||
| + | [http://www.sciencemuseum.org.uk/antenna/dnaorigami/index.asp The art of DNA origami] | ||
| + | [http://www.ablestable.com/resources/library/thecolumn/2006/037.htm Science and Creativity] | ||
| + | [http://www.azonano.com/details.asp?ArticleID=1466 From Nautilus to Nanobo(a)ts] | ||
| + | [http://fora.tv/fora/showthread.php?t=451 The Sims] | ||
| + | [http://www.uctv.tv/search-details.asp?showID=7153 A New Kind of Science] | ||
=== These people do great things === | === These people do great things === | ||
| Line 94: | Line 207: | ||
Albert Libchaber [http://www.rockefeller.edu/research/abstract.php?id=93] | Albert Libchaber [http://www.rockefeller.edu/research/abstract.php?id=93] | ||
Carlos Bustamante's lab [http://alice.berkeley.edu/] | Carlos Bustamante's lab [http://alice.berkeley.edu/] | ||
| + | Darko Stefanovic [http://www.cs.unm.edu/~darko/] | ||
David Deamer [http://www.chemistry.ucsc.edu/deamer_d.html] | David Deamer [http://www.chemistry.ucsc.edu/deamer_d.html] | ||
| + | David Liu [http://www.fas.harvard.edu/~biophys/David_R_Liu.htm] | ||
| + | Ehud Shapiro [http://www.wisdom.weizmann.ac.il/~udi/] | ||
Eric Kool’s group [http://www.stanford.edu/group/kool/] | Eric Kool’s group [http://www.stanford.edu/group/kool/] | ||
Erik Winfree [http://www.dna.caltech.edu/~winfree/] | Erik Winfree [http://www.dna.caltech.edu/~winfree/] | ||
Fred Menger’s group [http://www.chemistry.emory.edu/faculty/menger/index.html] | Fred Menger’s group [http://www.chemistry.emory.edu/faculty/menger/index.html] | ||
| + | Friedrich Simmel [http://www.nano.physik.uni-muenchen.de/nanobio/group/fritz.html] | ||
| + | Gerald Joyce [http://exobio.ucsd.edu/joyce.htm] | ||
| + | Hao Yan [http://www.biodesign.asu.edu/labs/yan/index.php] | ||
Jack Szostak’s lab [http://genetics.mgh.harvard.edu/szostakweb/] | Jack Szostak’s lab [http://genetics.mgh.harvard.edu/szostakweb/] | ||
| - | + | Len Adleman [http://www.usc.edu/dept/molecular-science/fm-adleman.htm] | |
| + | Luc Jaeger [http://www.cnsi.ucsb.edu/directory/faculty/jaeger/jaeger.html] | ||
| + | Molecular Computing Group [https://digamma.cs.unm.edu/wiki/bin/view/McogPublicWeb/WebHome] | ||
| + | Ned Seeman [http://seemanlab4.chem.nyu.edu/nanotech.html] | ||
| + | Peixuan Guo [http://www.vet.purdue.edu/PeixuanGuo/index.html] | ||
Pier Luigi Luisi’s group [http://www.plluisi.org/index.html] | Pier Luigi Luisi’s group [http://www.plluisi.org/index.html] | ||
| + | 'Protolife' by Norman Packard & Mark Bedau [http://www.protolife.net/] | ||
Radhika Nagpal [http://www.eecs.harvard.edu/~rad/] | Radhika Nagpal [http://www.eecs.harvard.edu/~rad/] | ||
| - | Steven | + | Richard Kiehl [http://www.ece.umn.edu/users/kiehl/index.html] |
| + | Seung-Wuk Lee [http://leelab.berkeley.edu/index_files/Page542.htm] | ||
| + | Steen Rasmussen [http://www.ees.lanl.gov/staff/steen/] | ||
| + | Steven Benner [http://www.chem.ufl.edu/benner.html] | ||
| + | Thomas LaBean [http://www.fitzpatrick.duke.edu/AboutCenter/faculty/about_labean.html] | ||
| + | William Shih [http://research2.dfci.harvard.edu/shih/] | ||
| + | Sorry whom aren’t on the list!<br> | ||
=== Local === | === Local === | ||
An Analysis of Synthetic Biology Research in Europe and North America [http://www2.spi.pt/synbiology/] | An Analysis of Synthetic Biology Research in Europe and North America [http://www2.spi.pt/synbiology/] | ||
| + | SB database [http://www.synthetic-biology.info/] | ||
DNA synthesis: ATG-Biosynthetics [http://www.atg-biosynthetics.com/] | DNA synthesis: ATG-Biosynthetics [http://www.atg-biosynthetics.com/] | ||
| + | febit [http://febit.de/europe/en/] | ||
| + | 'Spiegel' about iGEM competition on August 14, 2006 [http://www.spiegel.de/spiegel/0,1518,431399,00.html] | ||
| + | 'Neue Zürcher Zeitung' about SB on August 23, 2006 | ||
| + | [http://www.nzz.ch/2006/08/23/ft/articleEEA5E.html] | ||
| + | New GEM on the simiki wiki page [http://ernie.imtek.uni-freiburg.de/simiki/GeneticallyEngineeredMachines] | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | __NOTOC__ | ||
Latest revision as of 19:04, 4 January 2008
|
The team (all stars)
| Students | iGEM instructors | Faculty/staff |
|---|---|---|
|
The motto
|
|
The project: DNA Folding
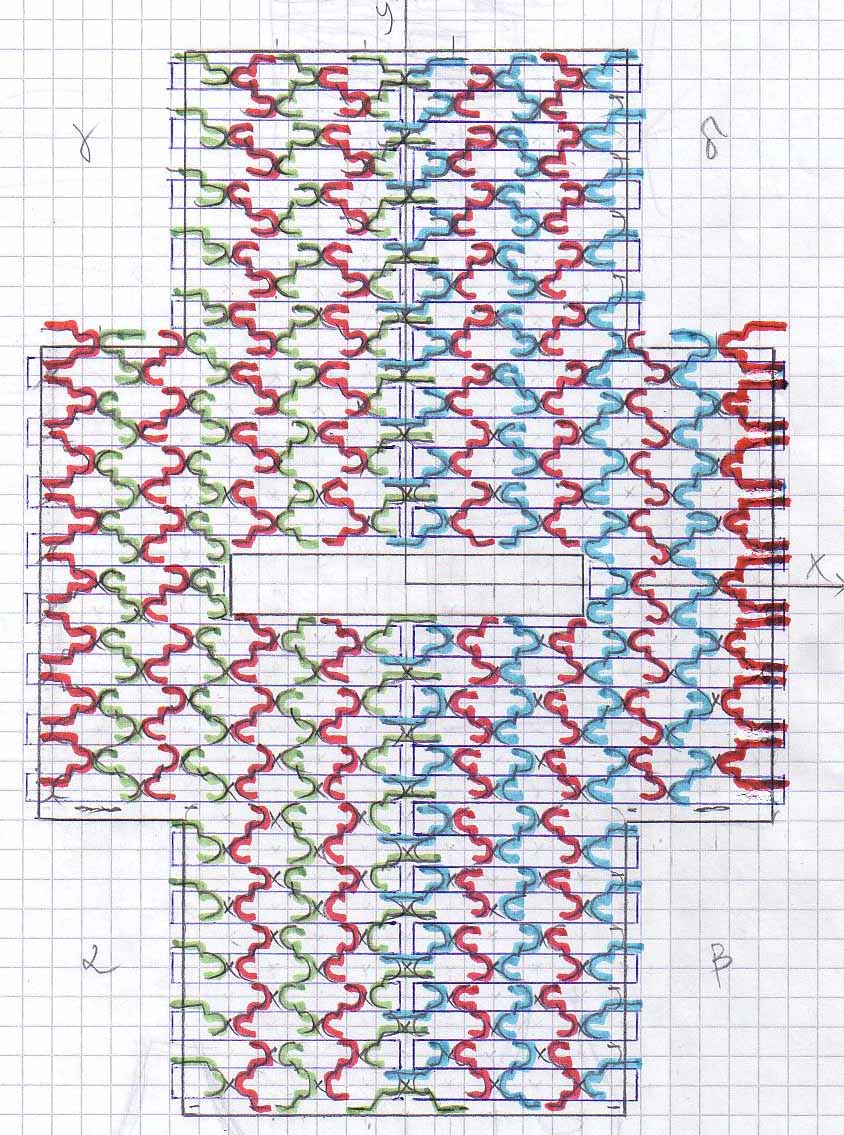
No.1 - The PipeThe idea is to design a strand of DNA such that it wraps into a specific shape. There are three different stages for this project: First, the DNA should fold into a two-dimensional rectangular sheet. Secondly, this sheet should wrap itself up into the shape of a short pipe. And last, these little pipes should hook themselves up to each other such that they form one single long pipe (more details).
No.2 - Haute Couture: Barbie NanoatelierOnce the process of DNA folding into 3D structures is understood, shapes can be chosen arbitrarily. The idea of the Barbie Nanoatelier is that the DNA should wrap into a 3D T-Shirt, 3D Pants, etc. "sea of parts" We need a library of molecular primitives that could be interchanged in the desirable way for a bottom-up design on the nanometer scale. Registry of DNA molecular parts and standardization is an actual question of bionanotechnology. We just put first examples of a new class of BioBricks - ORIGAMI BioBriks into [http://partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2006partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2006&group=Freiburg depository] and publish basic ideas [http://omnibus.uni-freiburg.de/~kouznet/DNA-plug.txt], [http://omnibus.uni-freiburg.de/~kouznet/T-shirt.txt], [http://omnibus.uni-freiburg.de/~kouznet/DNA-NanoSwarm.txt]. We try to merge a ‘dead DNA’ with an ‘alive DNA’. Surrealistic Science We also try to pump some aesthetic principles and rules (symmetry, periodic patterns, recursion, and plasticity) into our creatures. We like it because it needs a huge of imagination and really very difficult. Nobody can build a bra! - von Neumann's self-reproducing ... bra :))
Sketches - T-Shirt from DNAThe DNA sequence for the T-Shirt design can be found [http://omnibus.uni-freiburg.de/~kouznet/t-shirt/T-shirt_M13mp18_NEB_d.txt here], the picture is [http://omnibus.uni-freiburg.de/~kouznet/t-shirt/T-shirt.jpg here], and the staples are [http://omnibus.uni-freiburg.de/~kouznet/t-shirt/T-shirt_staples.txt here] there. Hey, venture capitalists, where are you? The price of all of 50 billion T-Shirts would be only 7200bases x €0.17/base = €1224. Don’t miss the chance. It could be a great business tomorrow! Ladies and Gentlemen, please, visit the demonstration of last Nike nano collection. (Enter) Oh, aroma of the podium [http://omnibus.uni-freiburg.de/~kouznet/LoveParade2005.mp3 music] Jobs: Still looking for catwalk models!!
No.3 - Individual projectsOther projects and prices could be found on individual Mutant's pages (Real iPod nano, Captain Nemo). In addition, you can find some hot information on ‘GEM Freiburg Fellow 2006’ page. The other toys will be unconventional computing, cryptography, nanoelectronics, nanooptics, drug delivery systems, smart nanomaterials, nanoswarm, eventually an artificial life and Origami Man. Time is money! |
GEM Freiburg
Club
SB Preliminary
|
Original ideas
Now we see it could be cheaper, but it will need a great intellectual impact. So, we’re opened for sponsors and relationships. CommentsAs I understand it, the idea is to create a "life automaton" consisting of 6 proteins? That is what I took out of your proposal. That seems quite ambitioned, but I can't really judge it, and even if it is quite ambitioned, that does not mean that it is bad, of course. My understanding says that you are using 6000 € for denovo synthesis of 6 genes required for making minimal life. Right? So, what if after all the simulations and tests on the computer, you order the DNA for minimal life and it turns out that it doesn't work... What will you do then? This is because biology is unpredictable and you must not assume that the DNA which you order will work in the first time itself... You must keep some backup money or plans... You have to separate specifically the general and specific objectives to long and short time; implications, obstacles and limitations. OK, fine.. So you want to go in a completely different direction by creating alternative life.. Good.. All the best for this project. Well, if you want to go to the extremes then are you limiting yourself to already established phenomena such as DNA, membranes, etc. You may want to adopt a completely new and novel ideas which might be simpler than DNA and other stuff of the normal cell. So, keep all the options open. Research the literature well and create Artificial Life... No more child labour, just molecule labour :) |
Hey Mutant, have a look!
The easy and serious way. Ja oder Nein?
DNA nanotechnology for newborn Mutants[http://www.npr.org/templates/story/story.php?storyId=5281562 Fun with DNA] [http://www.sciencemuseum.org.uk/antenna/dnaorigami/index.asp The art of DNA origami] [http://www.ablestable.com/resources/library/thecolumn/2006/037.htm Science and Creativity] [http://www.azonano.com/details.asp?ArticleID=1466 From Nautilus to Nanobo(a)ts] [http://fora.tv/fora/showthread.php?t=451 The Sims] [http://www.uctv.tv/search-details.asp?showID=7153 A New Kind of Science] These people do great thingsAlbert Libchaber [http://www.rockefeller.edu/research/abstract.php?id=93]
Carlos Bustamante's lab [http://alice.berkeley.edu/]
Darko Stefanovic [http://www.cs.unm.edu/~darko/]
David Deamer [http://www.chemistry.ucsc.edu/deamer_d.html]
David Liu [http://www.fas.harvard.edu/~biophys/David_R_Liu.htm]
Ehud Shapiro [http://www.wisdom.weizmann.ac.il/~udi/]
Eric Kool’s group [http://www.stanford.edu/group/kool/]
Erik Winfree [http://www.dna.caltech.edu/~winfree/]
Fred Menger’s group [http://www.chemistry.emory.edu/faculty/menger/index.html]
Friedrich Simmel [http://www.nano.physik.uni-muenchen.de/nanobio/group/fritz.html]
Gerald Joyce [http://exobio.ucsd.edu/joyce.htm]
Hao Yan [http://www.biodesign.asu.edu/labs/yan/index.php]
Jack Szostak’s lab [http://genetics.mgh.harvard.edu/szostakweb/]
Len Adleman [http://www.usc.edu/dept/molecular-science/fm-adleman.htm]
Luc Jaeger [http://www.cnsi.ucsb.edu/directory/faculty/jaeger/jaeger.html]
Molecular Computing Group [1]
Ned Seeman [http://seemanlab4.chem.nyu.edu/nanotech.html]
Peixuan Guo [http://www.vet.purdue.edu/PeixuanGuo/index.html]
Pier Luigi Luisi’s group [http://www.plluisi.org/index.html]
'Protolife' by Norman Packard & Mark Bedau [http://www.protolife.net/]
Radhika Nagpal [http://www.eecs.harvard.edu/~rad/]
Richard Kiehl [http://www.ece.umn.edu/users/kiehl/index.html]
Seung-Wuk Lee [http://leelab.berkeley.edu/index_files/Page542.htm]
Steen Rasmussen [http://www.ees.lanl.gov/staff/steen/]
Steven Benner [http://www.chem.ufl.edu/benner.html]
Thomas LaBean [http://www.fitzpatrick.duke.edu/AboutCenter/faculty/about_labean.html]
William Shih [http://research2.dfci.harvard.edu/shih/]
Sorry whom aren’t on the list! LocalAn Analysis of Synthetic Biology Research in Europe and North America [http://www2.spi.pt/synbiology/] SB database [http://www.synthetic-biology.info/] DNA synthesis: ATG-Biosynthetics [http://www.atg-biosynthetics.com/] febit [http://febit.de/europe/en/] 'Spiegel' about iGEM competition on August 14, 2006 [http://www.spiegel.de/spiegel/0,1518,431399,00.html] 'Neue Zürcher Zeitung' about SB on August 23, 2006 [http://www.nzz.ch/2006/08/23/ft/articleEEA5E.html] New GEM on the simiki wiki page [http://ernie.imtek.uni-freiburg.de/simiki/GeneticallyEngineeredMachines] |