ETH2006 xor
From 2006.igem.org
m |
(→Sensitivity analysis) |
||
| (38 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | →[[ETH Zurich 2006# | + | →[[ETH Zurich 2006#System modeling|back to main page]] |
== XOR gate == | == XOR gate == | ||
| - | The XOR gate's | + | The XOR gate's PoPS output activity should be correlated to the PoPS input activity as shown in the picture: |
input A ^ | input A ^ | ||
| Line 14: | Line 14: | ||
output: High, Low, Dont care | output: High, Low, Dont care | ||
| - | === | + | === Implementation === |
| - | ==== | + | ==== Implementation alternatives ==== |
| - | + | We came up with a number of concepts for the implementation of the XOR, initially without considering the existence such biological systems. Then, while our biologists started searching the literature, we modelled the variants to see which we might want to favor. | |
| - | + | Here we pick out two variants with corresponding simulation results to give an idea of the process of development. Note that at this stage, the models still contain minor inaccuracies such as missing ribosome binding sites, which we corrected later on. | |
{| | {| | ||
| Line 25: | Line 25: | ||
|} | |} | ||
| - | + | Further development of the first variant resulted in the current implementation of the XOR gate. | |
| - | ==== | + | ==== Current implementation ==== |
Part to be used in the prototype system: [http://partsregistry.org/Part:BBa_J34200 BBa_J34200] | Part to be used in the prototype system: [http://partsregistry.org/Part:BBa_J34200 BBa_J34200] | ||
| Line 34: | Line 34: | ||
Signal 1 leads to the expression of a bi-modular transcription factor where the DNA binding domain is separated from the transcriptional activation domain by a Tobacco Etch Virus (TEV) protease cleavage site. Moreover, Signal 1 triggers the expression of the highly sequence-specific Tobacco Vein Mottling Virus (TVMV) protease. On the other hand, Signal 2 leads to the expression of an almost identical transcription factor, but in this case the DNA binding domain is separated from the transcriptional activation domain by a TVMV protease cleavage site. In addition, Signal 2 induces the transcription of the TEV protease. Therefore, the presence of either one of the signals leads to a stable transcription factor, which can initiate the transcription of a downstream reporter, while the presence of both signals leads to the mutual cleavage of both transcription factors by the specific proteases. | Signal 1 leads to the expression of a bi-modular transcription factor where the DNA binding domain is separated from the transcriptional activation domain by a Tobacco Etch Virus (TEV) protease cleavage site. Moreover, Signal 1 triggers the expression of the highly sequence-specific Tobacco Vein Mottling Virus (TVMV) protease. On the other hand, Signal 2 leads to the expression of an almost identical transcription factor, but in this case the DNA binding domain is separated from the transcriptional activation domain by a TVMV protease cleavage site. In addition, Signal 2 induces the transcription of the TEV protease. Therefore, the presence of either one of the signals leads to a stable transcription factor, which can initiate the transcription of a downstream reporter, while the presence of both signals leads to the mutual cleavage of both transcription factors by the specific proteases. | ||
| - | Our bi-modular transcription factor consists of the DNA binding domain of the bacteriophage lambda repressor protein (λcI) fused to the N-terminal domain of the RNA polymerase α subunit. Binding of this transcription factor to its cognate promoter – the λ operator – initiates the transcription of a downstream reporter (in this case | + | Our bi-modular transcription factor consists of the DNA binding domain of the bacteriophage lambda repressor protein (λcI) fused to the N-terminal domain of the RNA polymerase α subunit. Binding of this transcription factor to its cognate promoter – the λ operator – initiates the transcription of a downstream reporter (in this case mCherry). |
The DNA binding domain (λcI) of the transcription factor is linked to N terminal domain of the α subunit of the RNA polymerase by a linker sequence consisting of three alanines, followed by the cognate protease cleavage sites and two alanines (AAA protease cleavage sequence AA). | The DNA binding domain (λcI) of the transcription factor is linked to N terminal domain of the α subunit of the RNA polymerase by a linker sequence consisting of three alanines, followed by the cognate protease cleavage sites and two alanines (AAA protease cleavage sequence AA). | ||
| Line 40: | Line 40: | ||
References: | References: | ||
| - | + | * Hu JC, Kornacker MG, Hochschild A. Escherichia coli one- and two-hybrid systems for the analysis and identification of protein-protein interactions. Methods. 2000 Jan;20(1):80-94. | |
| - | interactions. Methods. 2000 Jan;20(1):80-94. | + | * Nallamsetty S, Kapust RB, Tozser J, Cherry S, Tropea JE, Copeland TD, Waugh DS. Efficient site-specific processing of fusion proteins by tobacco vein mottling virus protease in vivo and in vitro. Protein Expr Purif. 2004 Nov;38(1):108-15. |
| - | + | * Kapust RB, Waugh DS. Controlled intracellular processing of fusion proteins by TEV protease. Protein Expr Purif. 2000 Jul;19(2):312-8. | |
| - | + | * Hochschild A., personal communication | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
[[Image:ETH xor.png|400px|center|current XOR gate implementation]] | [[Image:ETH xor.png|400px|center|current XOR gate implementation]] | ||
| - | === | + | === Modeling === |
| - | + | We are interested in the input/output behavior at steady-state, that is when the system has reached equilibra and the concentrations of concerned species don't change anymore. Since we have 2 inputs and 1 ouptut, simulation results will be illustrated as a surface plot, inputs on the x/y axes and output on the z axis. | |
| - | ==== | + | ==== Some Formulas ==== |
| - | + | (''general explanatory notes from'' [[Template:ETH_Modeling_Formulas|this template]]) | |
| + | {| style="background-color:#dddddd;border-spacing:10px;" | ||
| + | |- | ||
| + | | {{ETH_Modeling_Formulas}} | ||
| + | |} | ||
| + | |||
| + | ==== Ideal xor gate ==== | ||
| + | The expected input/output behavior of an ideal xor gate looks like this (better with less green and steeper flanks): | ||
{| | {| | ||
|- | |- | ||
| - | | align="center" | '' | + | | align="center" | ''top view'' || align="center" | ''3d surface plot'' |
|- | |- | ||
| [[Image:XOR_Ideal_Flat.png|200px|center|behavior of an ideal XOR gate, topside view]] | | [[Image:XOR_Ideal_Flat.png|200px|center|behavior of an ideal XOR gate, topside view]] | ||
| Line 67: | Line 69: | ||
| colspan="2" align="center" | ''behavior of an ideal XOR gate''<br/>(x/y: ''inputs'', z: ''output'') | | colspan="2" align="center" | ''behavior of an ideal XOR gate''<br/>(x/y: ''inputs'', z: ''output'') | ||
|} | |} | ||
| - | + | Note that the anterior low (blue) part has low inputs, the rear one has high inputs. | |
| - | ==== | + | ==== Early simulation results of variants ==== |
| - | + | The following figures show early simulation results of the implementation variants 1 and 2. Note that also the models contain inaccuracies and even defects at this stage. | |
{| | {| | ||
| Line 82: | Line 84: | ||
|} | |} | ||
| - | + | Both plots show behaviors far from that of the ideal XOR gate, but the first variant at least has high output if one or the other, but not both inputs are high. Further on, the first variant seemed to be less sensitive to parameter changes. In fact, it was very hard to find ''any'' parameter values for model 2 ''at all'' which showed XOR-like behavior at least in the broadest sense. | |
| - | + | Not only because of these results (biology also had a voice in this chapter), but definitely backing the decision, we went for variant 1 from here on. | |
| - | ==== | + | ==== Current simulation results ==== |
{| | {| | ||
|- | |- | ||
| - | | align="center" | '' | + | | align="center" | ''top view, ht<sub>Prot</sub>=2h'' || align="center" | ''3d surface plot, ht<sub>Prot</sub>=2h'' || align="center" | ''3d surface plot, ht<sub>Prot</sub>=5h'' |
|- | |- | ||
| [[image:ETH_Sim_Xor_2h_flat.png|300px|center|topside view of simulation results for XOR gate, protein halflife period: 2h]] | | [[image:ETH_Sim_Xor_2h_flat.png|300px|center|topside view of simulation results for XOR gate, protein halflife period: 2h]] | ||
| Line 98: | Line 100: | ||
|} | |} | ||
| - | + | For every plot, 25 (left) / 100 (middle, right) ODE simulations were performed, simulating a time period of 12h. Simulations with 2h / 5h halflife period for proteins have been run, showing little difference in the qualitative outcome, though. Complete parameter allocation can be found in the [[ETH_Simulation_Scripts|matlab scripts]] as well as the system of ordinary differential equations (ODEs). | |
| - | + | '''About input rates''' (''general explanatory notes from'' [[Template:ETH_Sim_Input_Rate|this template]]) | |
| - | + | {| style="background-color:#dddddd;" | |
| - | + | |- | |
| - | + | | {{ETH_Sim_Input_Rate}} | |
| - | + | |} | |
| - | |||
| - | |||
| - | |||
| - | |||
| - | we | + | In order to find out about other important, say ''sensitive'' parameters, we wanted to do this more systematically. The keyword has already been given: sensitivity analisys. |
| - | + | ==== Sensitivity analysis ==== | |
| + | '''Sensitivity matrices''' (''general explanatory notes from'' [[Template:ETH_Sensitivity_Matrices|this template]]) | ||
| + | {| style="background-color:#dddddd;border-spacing:10px;" | ||
| + | |- | ||
| + | | {{ETH_Sensitivity_Matrices}} | ||
| + | |} | ||
| - | |||
| + | We computed sensitivity matrices for different input combinations. The results for the XOR gate are illustrated in the following bar3 plot: | ||
{| | {| | ||
|- | |- | ||
| - | | align="center" | ''XOR: | + | | align="center" | ''XOR: Sensitivity Analysis'' |
|- | |- | ||
| - | | [[image:ETH_Sens_XOR_5h.jpg|600px|center| | + | | <!--[[image:ETH_Sens_XOR_5h.jpg|600px|center|Normalized Sensitivity of the parameters for XOR gate]]--> [[image:ETH_Sens_XOR_5h.png|700px|center|Normalized Sensitivity of the parameters for XOR gate]] |
|- | |- | ||
| - | | colspan="2" align="center" | '' | + | | colspan="2" align="center" | ''Normalized Sensitivity of the parameters for XOR gate'' |
|} | |} | ||
| - | + | The z axis shows the absolute normalized sensitivities, the other axes correspond to different input combinations (shorter axis) and the parameters. The left most series show inputs, not sensitivities (3x3 inputs, each <tt>[low, medium, high]</tt>). The unlabeled series is a placeholder for aesthetic purposes only. Detailed explanation of the parameter abbreviations can be found [[ETH_Sim_Constants|here]]. Parameters are postfixed with the state variable they belong to: | |
| + | I1/I2 : Transcription factors TF1/TF2 | ||
| + | I1p/I2p : Proteases P1/P2 | ||
| + | I1inact/I2inact : Unfunctional cleaved transcription factors T+F1 / T+F2 | ||
| + | Out : Reporter protein, here RFP (mCherry) | ||
| + | mOut : mRNA for Out | ||
| - | + | The figure shows | |
| + | * Insensitivity with respect to the parameters <tt>d-I1inact/d-I2inact</tt>, and indeed degredation rate of the unfunctional cleaved transcription factors has no influence on the system ouput | ||
| + | * No sensitivity to various <tt>k</tt> parameters involved in the cleavage reactions if either one or the other input is 0. This makes sense since 0 rate on one input means no action on both cleavage reactions, and thus the concerned parameters just don't matter. | ||
| + | * Relatively high sensitivity to changes in the input rates, which is actually what we want! | ||
| + | * High sensifivity regarding the <tt>k-I1 / k-I2</tt> values for the transcription factors, and lower sensivitity for <tt>k-I1p / k-I2p</tt>. These <tt>k</tt> values act as amplification factors for protein synthesis, and indeed it is important that transcription factor concentration is high enough, where already little protease concentrations are sufficient. By the way, we can affect these <tt>k</tt> values by using ribosome binding sites of different strengths. | ||
| + | * ... | ||
| - | === | + | ==== More (old) simulation results ==== |
| - | + | [[ETH_Sim_Mod_Xor|simulation results]] / [[ETH_Sens_Xor|sensitivity analysis]] | |
| - | + | ||
| - | + | ||
| - | === | + | === Parts === |
[http://partsregistry.org/Part:BBa_J34200 BBa_J34200] | [http://partsregistry.org/Part:BBa_J34200 BBa_J34200] | ||
| - | →[[ETH Zurich 2006# | + | →[[ETH Zurich 2006#System modeling|back to main page]] |
Latest revision as of 13:26, 1 November 2006
Contents |
XOR gate
The XOR gate's PoPS output activity should be correlated to the PoPS input activity as shown in the picture:
input A ^
| H D L
| D D D
| L D H
+--------->
input B
output: High, Low, Dont care
Implementation
Implementation alternatives
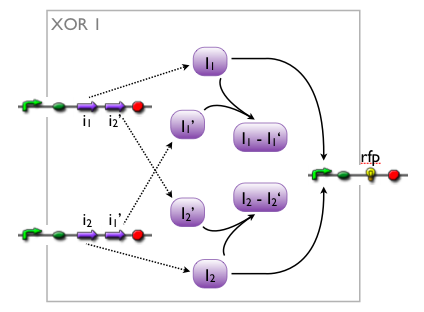
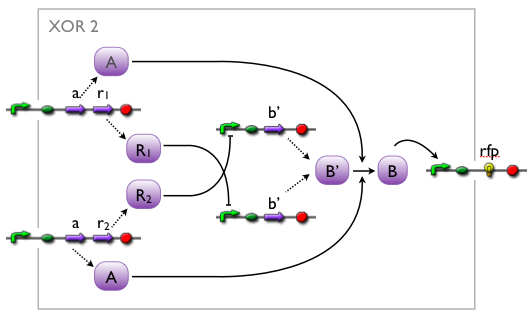
We came up with a number of concepts for the implementation of the XOR, initially without considering the existence such biological systems. Then, while our biologists started searching the literature, we modelled the variants to see which we might want to favor.
Here we pick out two variants with corresponding simulation results to give an idea of the process of development. Note that at this stage, the models still contain minor inaccuracies such as missing ribosome binding sites, which we corrected later on.
Further development of the first variant resulted in the current implementation of the XOR gate.
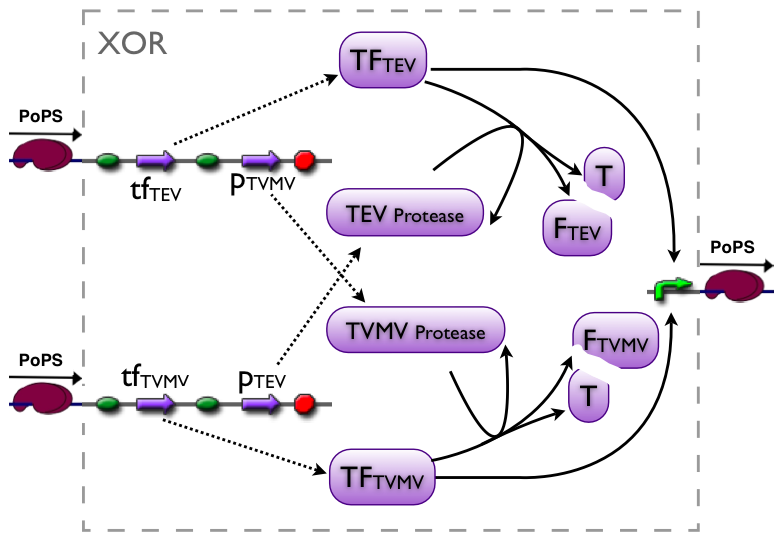
Current implementation
Part to be used in the prototype system: [http://partsregistry.org/Part:BBa_J34200 BBa_J34200]
An XOR gate produces an output only if one of the two possible input signals is present. In our implementation the gene expression triggered by Signal 1 inhibits the gene expression triggered by Signal 2 and vice versa.
Signal 1 leads to the expression of a bi-modular transcription factor where the DNA binding domain is separated from the transcriptional activation domain by a Tobacco Etch Virus (TEV) protease cleavage site. Moreover, Signal 1 triggers the expression of the highly sequence-specific Tobacco Vein Mottling Virus (TVMV) protease. On the other hand, Signal 2 leads to the expression of an almost identical transcription factor, but in this case the DNA binding domain is separated from the transcriptional activation domain by a TVMV protease cleavage site. In addition, Signal 2 induces the transcription of the TEV protease. Therefore, the presence of either one of the signals leads to a stable transcription factor, which can initiate the transcription of a downstream reporter, while the presence of both signals leads to the mutual cleavage of both transcription factors by the specific proteases.
Our bi-modular transcription factor consists of the DNA binding domain of the bacteriophage lambda repressor protein (λcI) fused to the N-terminal domain of the RNA polymerase α subunit. Binding of this transcription factor to its cognate promoter – the λ operator – initiates the transcription of a downstream reporter (in this case mCherry).
The DNA binding domain (λcI) of the transcription factor is linked to N terminal domain of the α subunit of the RNA polymerase by a linker sequence consisting of three alanines, followed by the cognate protease cleavage sites and two alanines (AAA protease cleavage sequence AA).
References:
- Hu JC, Kornacker MG, Hochschild A. Escherichia coli one- and two-hybrid systems for the analysis and identification of protein-protein interactions. Methods. 2000 Jan;20(1):80-94.
- Nallamsetty S, Kapust RB, Tozser J, Cherry S, Tropea JE, Copeland TD, Waugh DS. Efficient site-specific processing of fusion proteins by tobacco vein mottling virus protease in vivo and in vitro. Protein Expr Purif. 2004 Nov;38(1):108-15.
- Kapust RB, Waugh DS. Controlled intracellular processing of fusion proteins by TEV protease. Protein Expr Purif. 2000 Jul;19(2):312-8.
- Hochschild A., personal communication
Modeling
We are interested in the input/output behavior at steady-state, that is when the system has reached equilibra and the concentrations of concerned species don't change anymore. Since we have 2 inputs and 1 ouptut, simulation results will be illustrated as a surface plot, inputs on the x/y axes and output on the z axis.
Some Formulas
(general explanatory notes from this template)
| To get our models into a form which can be simulated, we needed to transform the wiring diagrams into a set of ODEs (ordinary differential equations), which in our case will be non-linear.
For every concerned species X, we write d[X]/dt = production - consumption For enzymatic transformation of substrate S into product P (catalyzed by enzyme E), we write k+1 k2
S + E <==> E•S --> P + E
k−1
d[S]/dt = −k+1[S][E] + k−1[E•S] − dS[S]
d[E]/dt = −k+1[S][E] + k−1[E•S] + k2[E•S] − dE[E]
d[E•S]/dt = k+1[S][E] − k−1[E•S] − k2[E•S] − dES[E•S]
d[P]/dt = + k2[E•S] − dP[P]
[S] : substrate concentration
[E] : enzyme conc.
[E•S] : concentration of enzyme-substrate complex
[P] : product concentration
kinetic constants:
kk+1 : building enzyme-substrate complex (forward)
kk−1 : resolving enzyme-substrate complex (backward)
k2 : product formation
dXXX : degradation constants
For constitutive transcription, we have constant production rate and simply write d[M]/dt = ktr•u − dM[M] [M] : mRNA concentration ktr : kinetic constant (transcription) u : system input, e.g. transcription rate (≅PoPS) dM : degradation constant for mRNA A transcriptional regulatory module can be described by and ODE of the following form: 1
d[M]/dt = ktr ( a + −−−−−−−−−−−−−− ) − dM[M]
1 + (K/[S])α•n
[M] : mRNA concentration
ktr : kinetic constant (transcription)
a : constitutive portion, 0 ≤ a < 1
[S] : inducer (α=+1) / repressor (α=−1) concentration
K : hill constant
n : hill coefficient
α : α=+1 for induction, α=−1 for repression
dM : degradation constant for mRNA
Finally, translation is usually modeled like this: d[P]/dt = ktl[M] − dP[P] [P] : product (protein) concentration [M] : mRNA concentration ktl : kinetic constant (translation) dP : degradation constant for protein P References:
|
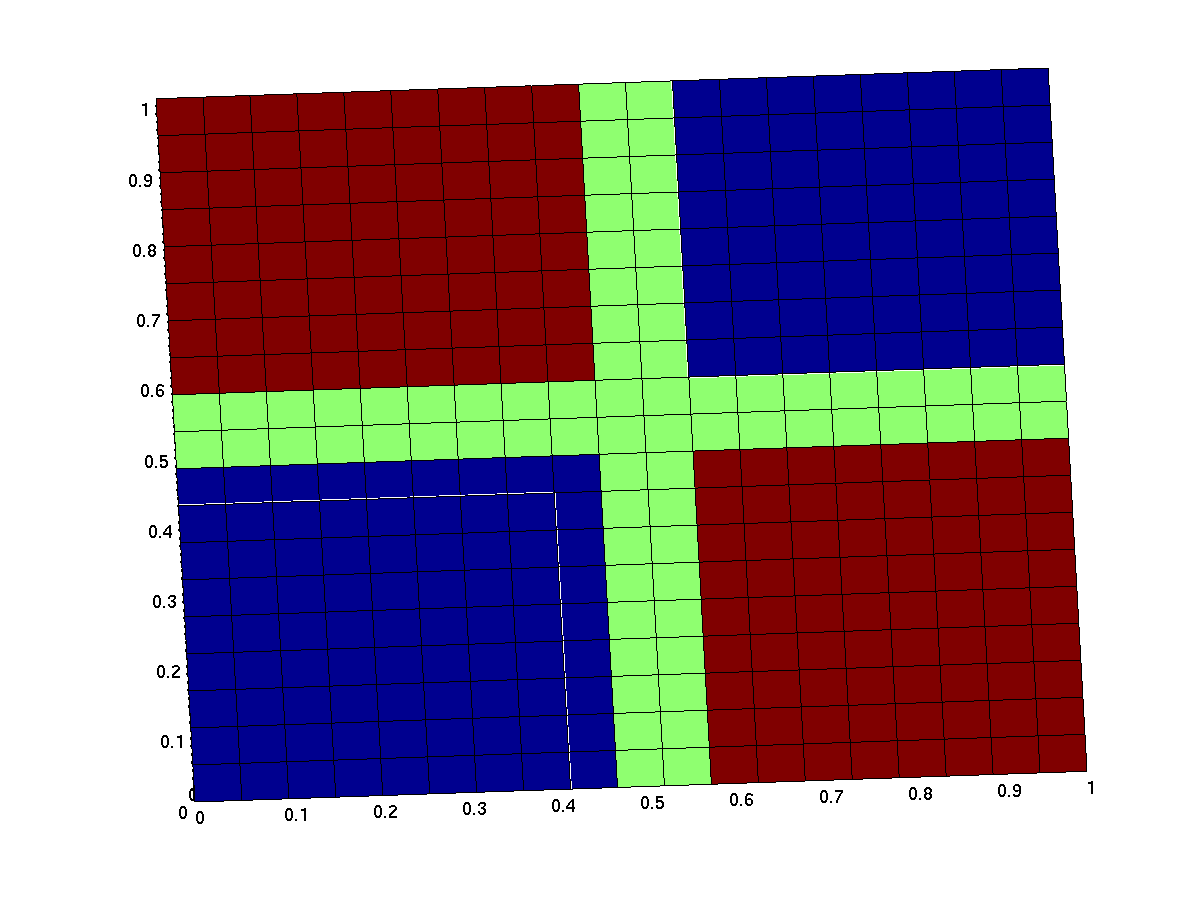
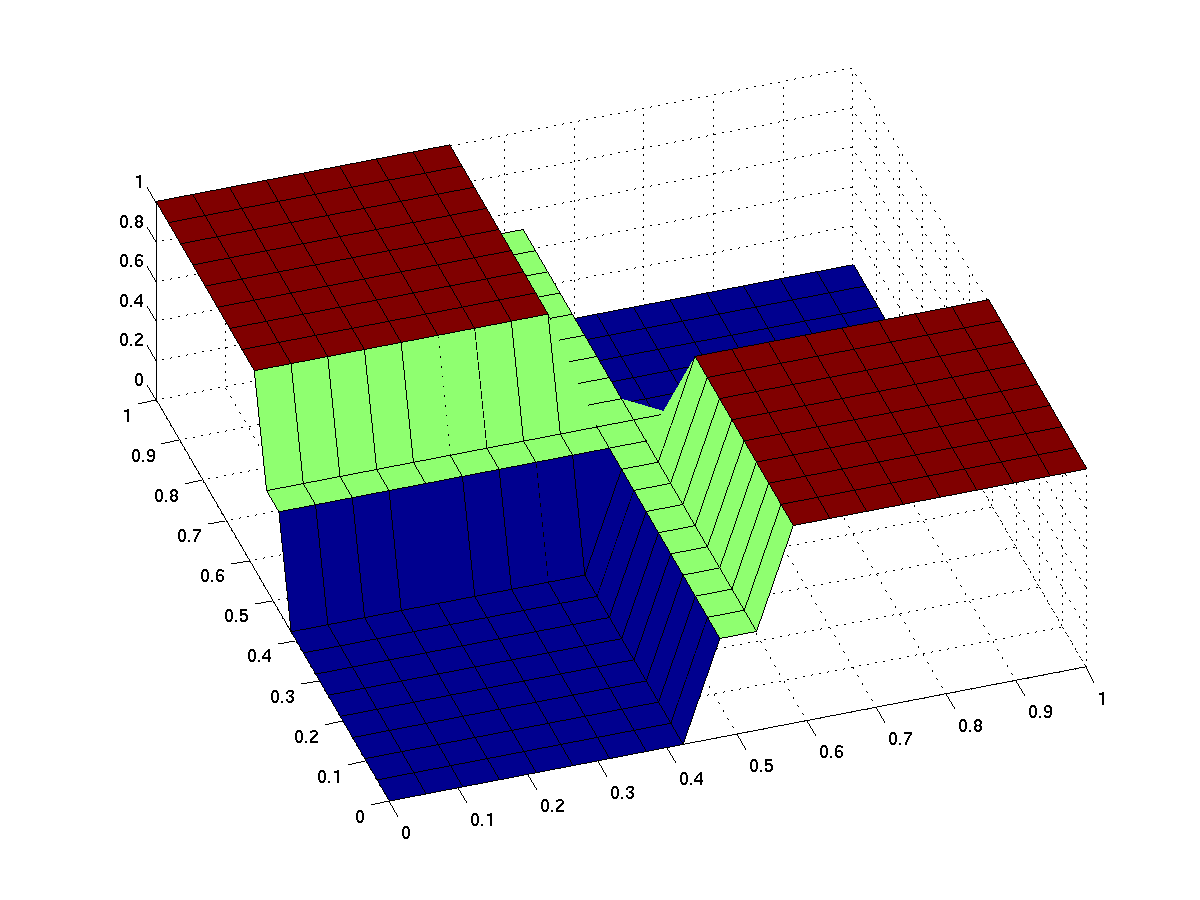
Ideal xor gate
The expected input/output behavior of an ideal xor gate looks like this (better with less green and steeper flanks):
| top view | 3d surface plot |
| behavior of an ideal XOR gate (x/y: inputs, z: output) | |
Note that the anterior low (blue) part has low inputs, the rear one has high inputs.
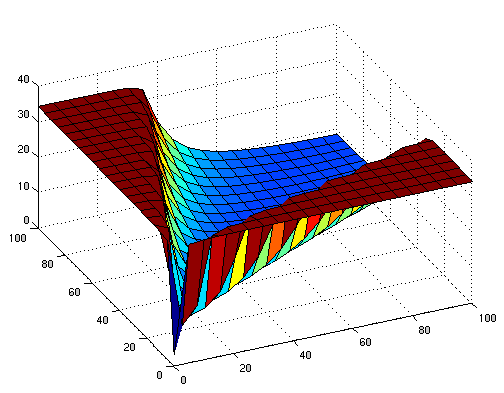
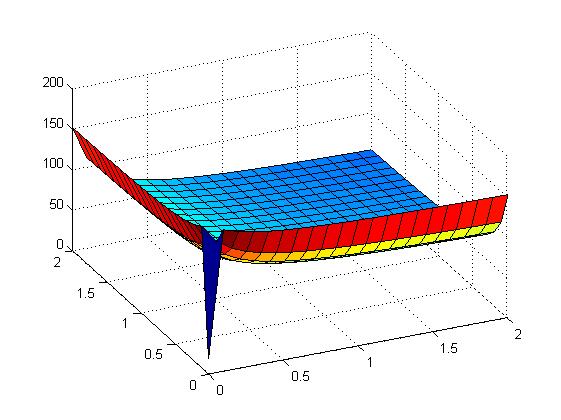
Early simulation results of variants
The following figures show early simulation results of the implementation variants 1 and 2. Note that also the models contain inaccuracies and even defects at this stage.
| Variant 1 | Variant 2 |
| surface plot of simulation results for XOR variants 1 and 2 (x/y: inputs, z: output) | |
Both plots show behaviors far from that of the ideal XOR gate, but the first variant at least has high output if one or the other, but not both inputs are high. Further on, the first variant seemed to be less sensitive to parameter changes. In fact, it was very hard to find any parameter values for model 2 at all which showed XOR-like behavior at least in the broadest sense.
Not only because of these results (biology also had a voice in this chapter), but definitely backing the decision, we went for variant 1 from here on.
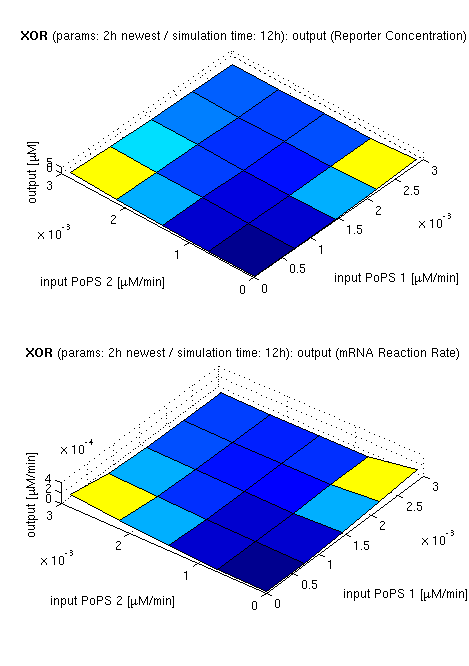
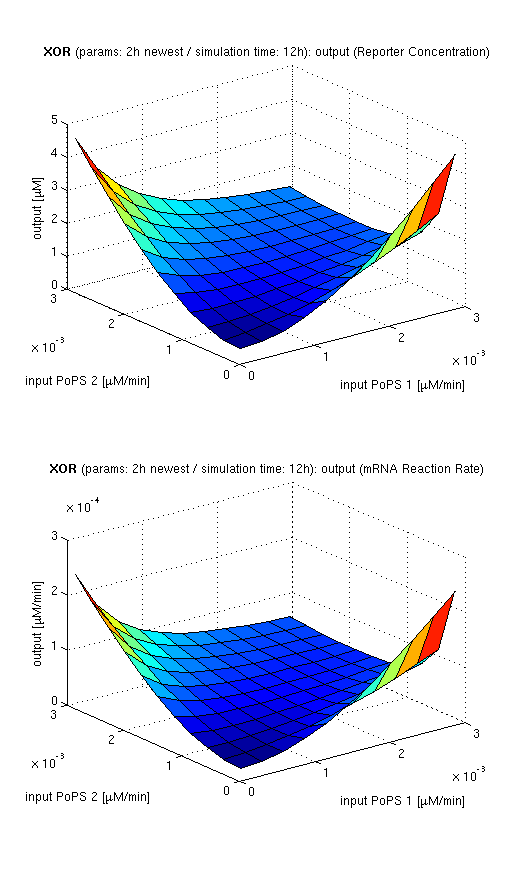
Current simulation results
| top view, htProt=2h | 3d surface plot, htProt=2h | 3d surface plot, htProt=5h |
| simulation results for XOR gate, different protein halflife periods top / bottom: output (z axis) is mRNA rate (∼PoPS) / reporter concentration (x/y: inputs, z: output, htProt: protein halflife period) | ||
For every plot, 25 (left) / 100 (middle, right) ODE simulations were performed, simulating a time period of 12h. Simulations with 2h / 5h halflife period for proteins have been run, showing little difference in the qualitative outcome, though. Complete parameter allocation can be found in the matlab scripts as well as the system of ordinary differential equations (ODEs).
About input rates (general explanatory notes from this template)
The input rates (PoPS) have been chosen in the range of mRNA transcription rate, which was estimated according to the following assumptions:
Amplifying/damping the input rates by small constant factors has influence on the qualitative outcome of the simulation.
We accounted for this by adding restriction enzyme sites to the DNA, so that we can cut out current RBS to replace it by another of different strength. |
In order to find out about other important, say sensitive parameters, we wanted to do this more systematically. The keyword has already been given: sensitivity analisys.
Sensitivity analysis
Sensitivity matrices (general explanatory notes from this template)
To analyze the sensitivity of the system for all concerned parameters, we compute the sensitivity matrix S:
S = (∂x/x) / (∂p/p) = (∂x/∂p) * (p/x) S : sensitivity matrix, #x rows, #p columns x : states (concentrations) p : parameters We use jacobian matrices of the system equations to compute the sensitivity matrix S. We therefore augment the set of differential equations by dS(t)/dt = Jx(t) S(t) + Jp(t) Jx(t) = ∂f(x,p,t)/∂x Jp(t) = ∂f(x,p,t)/∂p Since we are interested in the output sensitivities at steady state, we set dS(t)/dt = 0 and can solve for S: S = − Jx \ Jp \ : matrix left division |
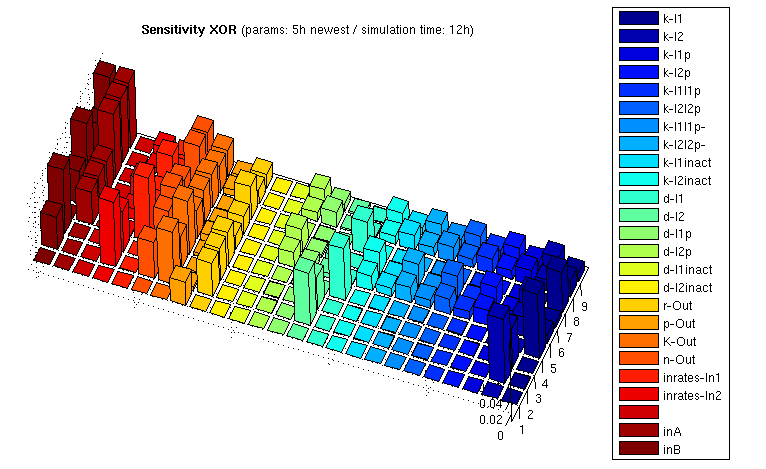
We computed sensitivity matrices for different input combinations. The results for the XOR gate are illustrated in the following bar3 plot:
| XOR: Sensitivity Analysis | |
| Normalized Sensitivity of the parameters for XOR gate | |
The z axis shows the absolute normalized sensitivities, the other axes correspond to different input combinations (shorter axis) and the parameters. The left most series show inputs, not sensitivities (3x3 inputs, each [low, medium, high]). The unlabeled series is a placeholder for aesthetic purposes only. Detailed explanation of the parameter abbreviations can be found here. Parameters are postfixed with the state variable they belong to:
I1/I2 : Transcription factors TF1/TF2 I1p/I2p : Proteases P1/P2 I1inact/I2inact : Unfunctional cleaved transcription factors T+F1 / T+F2 Out : Reporter protein, here RFP (mCherry) mOut : mRNA for Out
The figure shows
- Insensitivity with respect to the parameters d-I1inact/d-I2inact, and indeed degredation rate of the unfunctional cleaved transcription factors has no influence on the system ouput
- No sensitivity to various k parameters involved in the cleavage reactions if either one or the other input is 0. This makes sense since 0 rate on one input means no action on both cleavage reactions, and thus the concerned parameters just don't matter.
- Relatively high sensitivity to changes in the input rates, which is actually what we want!
- High sensifivity regarding the k-I1 / k-I2 values for the transcription factors, and lower sensivitity for k-I1p / k-I2p. These k values act as amplification factors for protein synthesis, and indeed it is important that transcription factor concentration is high enough, where already little protease concentrations are sufficient. By the way, we can affect these k values by using ribosome binding sites of different strengths.
- ...
More (old) simulation results
simulation results / sensitivity analysis
Parts
[http://partsregistry.org/Part:BBa_J34200 BBa_J34200]