Freiburg University 2006
From 2006.igem.org
Kouznetsov (Talk | contribs) |
Kouznetsov (Talk | contribs) |
||
| Line 56: | Line 56: | ||
==== Individual projects ==== | ==== Individual projects ==== | ||
| - | Other projects and prices could be found on individual Mutant's pages. In addition, you can find some hot information on ‘GEM Freiburg Fellow 2006’ page. The other toys will be unconventional computing, nanoelectronics, nanooptics, drug delivery systems, smart nanomaterials, nanoswarm, and eventually an | + | Other projects and prices could be found on individual Mutant's pages. In addition, you can find some hot information on ‘GEM Freiburg Fellow 2006’ page. The other toys will be unconventional computing, nanoelectronics, nanooptics, drug delivery systems, smart nanomaterials, nanoswarm, and eventually an artificial life. Time is money! |
== GEM Freiburg == | == GEM Freiburg == | ||
Revision as of 08:04, 4 October 2006
This term was invented by Martin Schneider on the Rule 110 Winter Workshop in 2004 [http://www.rule110.org/amhso/index.html]. We play without rules. We discover the rules that govern life, the universe and everything to exploit these rules and to create Artificial Life. Our short-time aim is the trip to Boston in November 2006 to take a prize in the iGEM.
The Team
Students
iGEM instructor
Faculty/staff
The project: DNA Folding
Basic Idea
Abstract
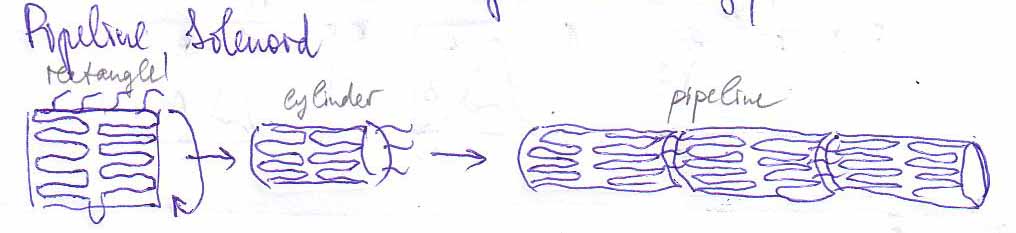
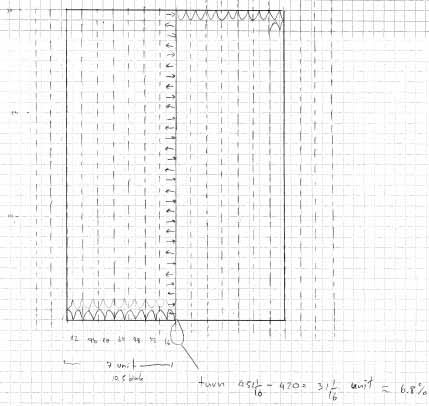
The idea is to design a strand of DNA such that it wraps into some meaningful shape. There are three different stages for this project: First, the DNA should fold into a two-dimensional rectangular sheet. Secondly, this sheet should wrap itself up into the shape of a short pipe. And last, these little pipes should hook themselves up to each other such that they form one single long pipe.
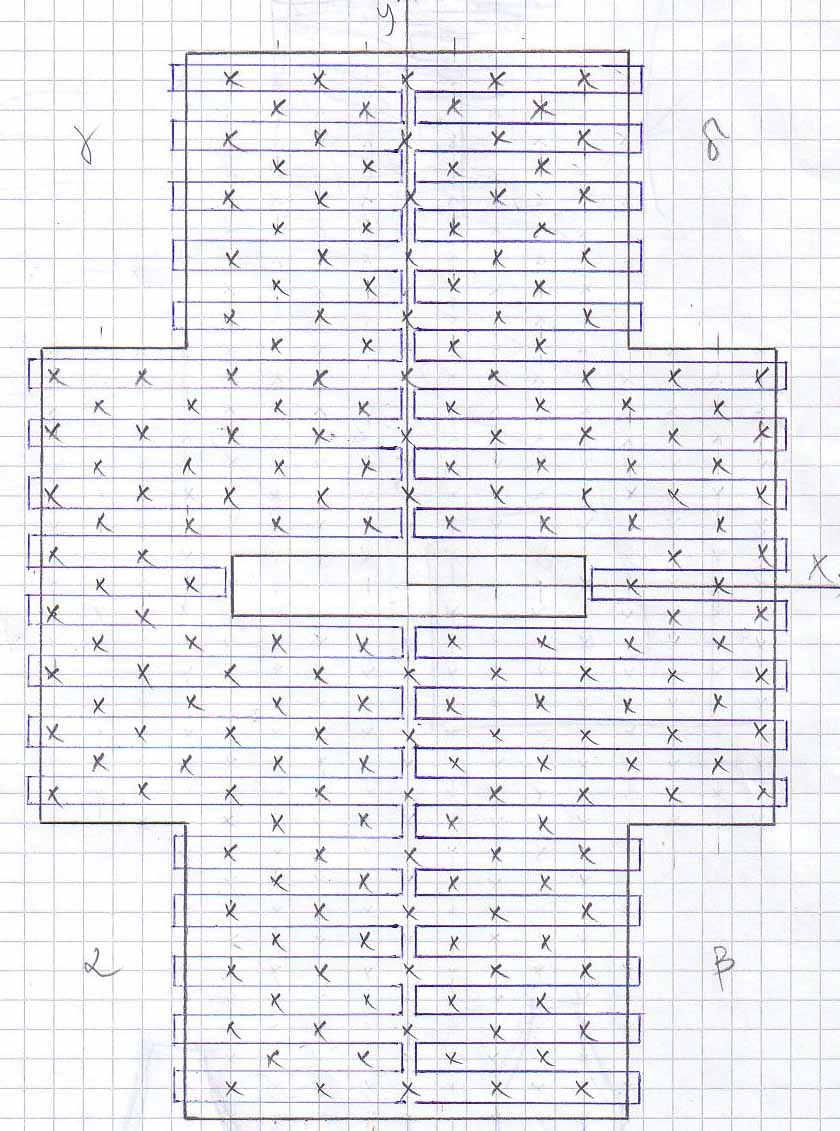
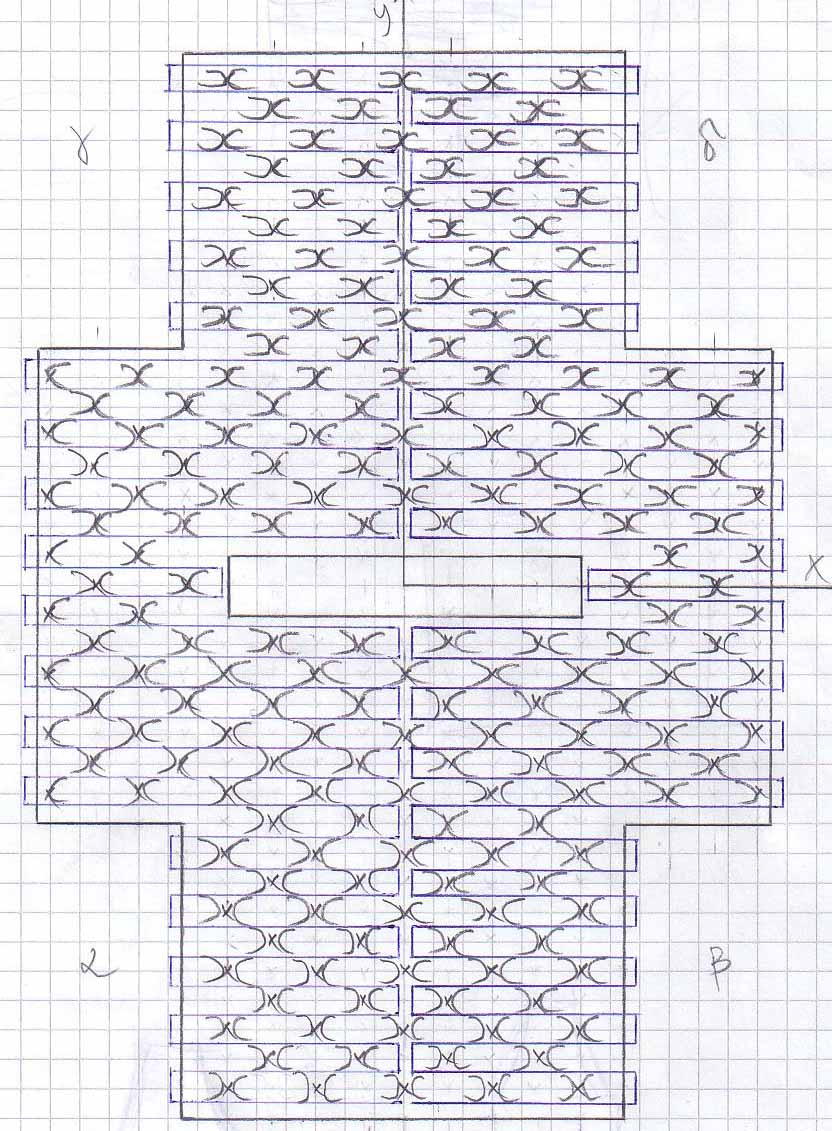
Scetches - Pipes
The DNA sequence for the Pipes design can be found [http://omnibus.uni-freiburg.de/~kouznet/pipe/Tube-M13mp18-NEB-d.txt here].
Application: Barbie Nanoatelier
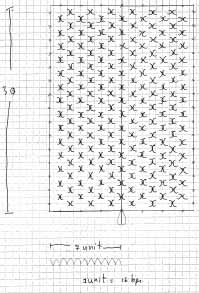
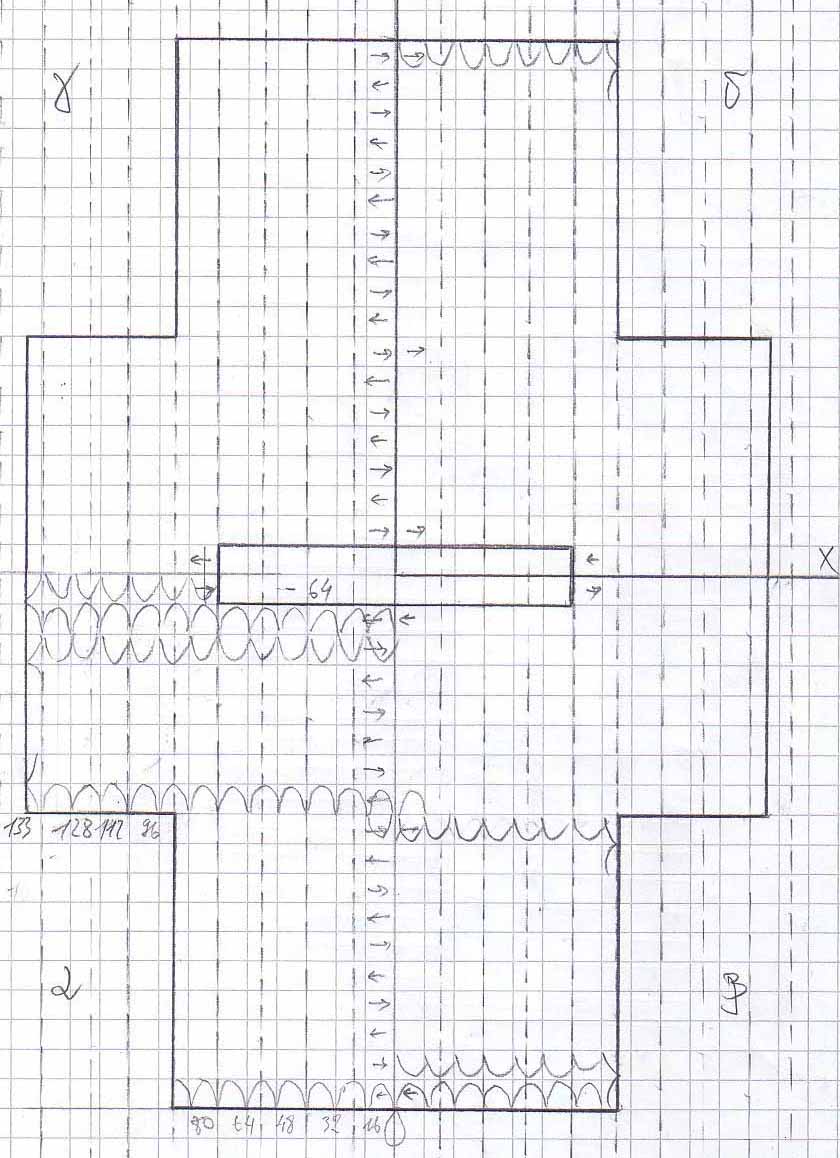
Once the process of DNA folding into 3D structures is understood, shapes can be chosen arbitrarily. The idea of the Barbie Nanoatelier is that the DNA should wrap into a 3D T-Shirt, 3D Pants, etc.
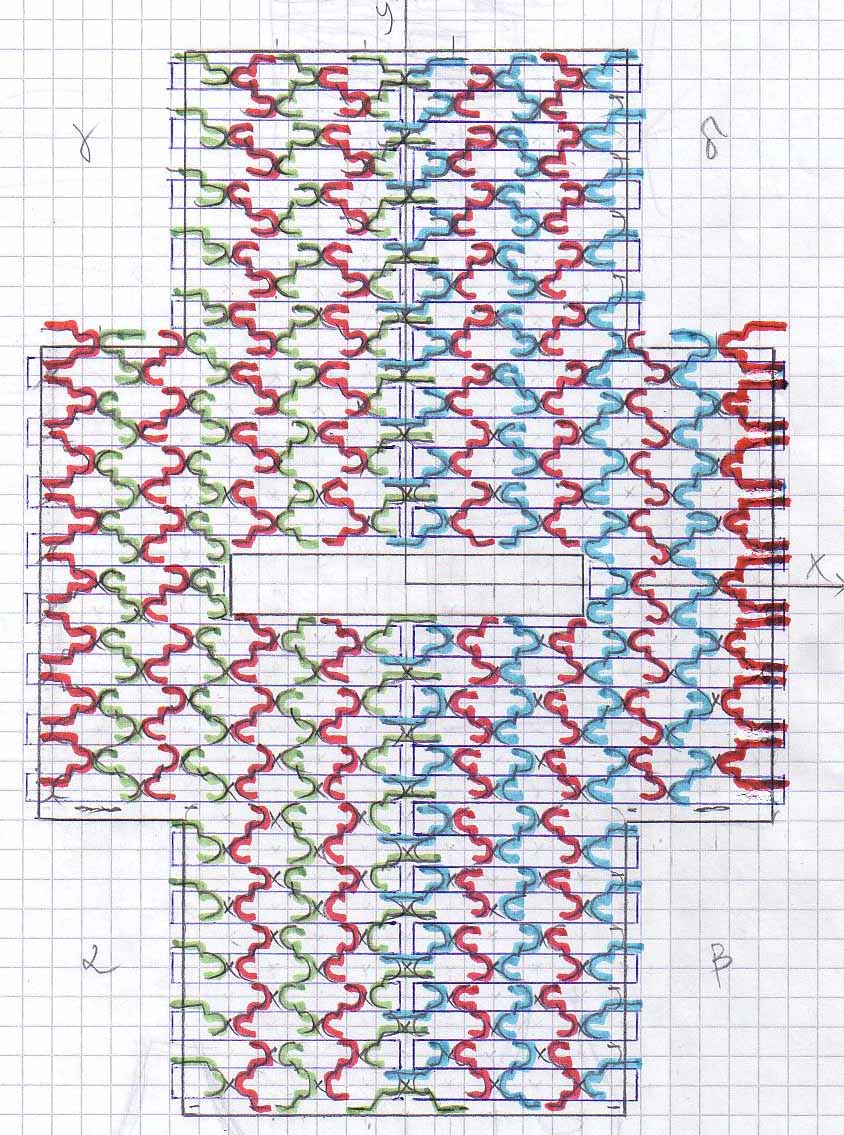
Scetches - T-Shirt
The DNA sequence for the T-Shirt design can be found [http://omnibus.uni-freiburg.de/~kouznet/t-shirt/T-shirt_M13mp18_NEB_d.txt here], the picture is [http://omnibus.uni-freiburg.de/~kouznet/t-shirt/T-shirt.jpg here], and the staples are [http://omnibus.uni-freiburg.de/~kouznet/t-shirt/T-shirt_staples.txt here] there.
Hey, venture capitalists, where are you? The price of all of 50 billion T-Shirts would be only 7200bases x €0.17/base = €1224. Don’t miss the chance. It could be a great business tomorrow!
Individual projects
Other projects and prices could be found on individual Mutant's pages. In addition, you can find some hot information on ‘GEM Freiburg Fellow 2006’ page. The other toys will be unconventional computing, nanoelectronics, nanooptics, drug delivery systems, smart nanomaterials, nanoswarm, and eventually an artificial life. Time is money!
GEM Freiburg
Club
- [http://omnibus.uni-freiburg.de/~kouznet/iGEM.htm Manifesto, Self-education program]
- [http://omnibus.uni-freiburg.de/~kouznet/Fellow06.htm GEM Freiburg Fellow 2006]
- [http://omnibus.uni-freiburg.de/~kouznet/GEMnews.htm News]
SB Preliminary
- DNA plug-and-play platform [http://omnibus.uni-freiburg.de/~kouznet/DNAplatform6.pdf 24.05.06] [http://omnibus.uni-freiburg.de/~kouznet/DNAplatform6-1.pdf 27.06.06]
- [http://omnibus.uni-freiburg.de/~kouznet/BioVLSI.pdf Very Large-Scale Integration design in Biology]
- [http://omnibus.uni-freiburg.de/~kouznet/GS-design.pdf Genetic systems design from the DNA modules]
Original idea
Hey Mutant, have a look!
The easy and serious way
- [http://www.edge.org/documents/archive/edge187.html#church George Church CONSTRUCTIVE BIOLOGY] - a nice paper about the challenges of modern biology
- [http://webcast.berkeley.edu/events/details.php?webcastid=15766 The webcasts from SB2.0] [http://openwetware.org/wiki/Synthetic_Biology:Synthetic_Biology_1.0/Videos and SB1.0]
These people do great things
Albert Libchaber [http://www.rockefeller.edu/research/abstract.php?id=93]
Carlos Bustamante's lab [http://alice.berkeley.edu/]
David Deamer [http://www.chemistry.ucsc.edu/deamer_d.html]
Eric Kool’s group [http://www.stanford.edu/group/kool/]
Erik Winfree [http://www.dna.caltech.edu/~winfree/]
Fred Menger’s group [http://www.chemistry.emory.edu/faculty/menger/index.html]
Jack Szostak’s lab [http://genetics.mgh.harvard.edu/szostakweb/]
Norman Packard’s Protolife [http://www.protolife.net/]
Pier Luigi Luisi’s group [http://www.plluisi.org/index.html]
Radhika Nagpal [http://www.eecs.harvard.edu/~rad/]
Steven Benner’s group [http://www.chem.ufl.edu/groups/benner/]
Local
An Analysis of Synthetic Biology Research in Europe and North America [http://www2.spi.pt/synbiology/] DNA synthesis: ATG-Biosynthetics [http://www.atg-biosynthetics.com/]