Predator-Prey Behavior
From 2006.igem.org
Back to ETH Zurich main page.
Back to Development Group.
Contents |
Intro and Principle
A predator-prey population dynamics should be mimicked by two similar (but not identical) bacterial populations which will "hunt" each other periodically. This will be visually detectable as an obscillating change in fluorescence from green to red, similar to a traffic light...
The "Mutual Killer" Approach
The two populations will be periodically exchanging the roles of predator and prey , by mutually activating a toxic gene. Timing should be controlled by concentration levels of inducers and, obviously, of the toxin.
The features
Population Red:
- constitutively expresses RFP.
- gene A: coding for quorum sensing factor A (e.g. AHL). Expression induced through external stimulus + auto-induction.
- gene toxR: encoding a toxic protein (ccdB?). Expression induced by C.
- gene C: coding for transcriptional activator C. Expression indirectly induced by B (activates phosphorelay).
Population Green:
- constitutively expresses GFP.
- gene B: encoding transcriptional activator B, constitutively expressed.
- gene toxG: encoding a toxic protein (ccdB?). Expression induced by a threshold concentration of A.
The Concept
The predator-prey behavior will be elicited by an external stimulus (e.g. IPTG or wathever), which will activate the transcription of the quorum sensing factor A in the red population. As soon as a threshold concentration of A is achieved, this will trigger transcription of a toxG gene in the green population. The tox gene encodes for a toxin,which will kill green cells to some extent (they should not all be killed). Moreover, the green population was constitutively expressing B, which can not cross the membrane unless the cells are dead... Therefore, death of green cells will lead to the release of B, which interacts with a receptor on the red population cells and induces a phosphorelay that leads to activation of expression of C, which in turn will induce transcription of the toxR gene. Then will cell density of the red population decrease, and a lower concentration of A will be present, so that the green survivors can begin to re-grow, etc etc etc etc etc etc.
Challenges
- Quite complicated, will it possibly work???
- Which toxin can be used to lower the population OD/cell count without killing the whole pop? Could CcdB be useful for this task?
- Will populations actually oscillate? Or will they soon reach a steady state?
- Literature shows that, after a while, prey population acquire resistance to the predator. Would that be possible in this case? Since populations are both prey and predator, would this plastic adaptation end up in a "draw"?
- Will bistability be a problem?
The "Nutrition-based" Approach
Introduction
The above solution is not really a predator-prey behavior (at least with the current intepretation) since they just kill each other but don't actually depend on each other in terms of feeding behavior. A suggestion that came up in the team was to make the prey population produce some real nutrition, e.g. amino acids, that the predator will depend on. So if the predator population exploits the prey population too much (by actually killing them, e.g. causing apoptosis which will release the nutrition stored in that prey-cell), there will be a lack of nutrition and the predator population will also shrink. This would create some real predator-prey dynamics.
The Features
Population Red:
- constitutively expresses RFP.
- gene A: coding for quorum sensing factor A (e.g. AHL). Expression induced through external stimulus + auto-induction.
- B auxotrophic (B could be e.g. an amino acid).
Population Green:
- constitutively expresses GFP.
- upregulated B biosynthesis and secretion.
- gene toxG: encoding a toxic protein (ccdB?). Expression induced by a threshold concentration of A.
The Concept
The predator-prey behavior will be elicited by an external stimulus (e.g. IPTG or wathever), which will activate the transcription of the quorum sensing factor A in the red population. As soon as a threshold concentration of A is achieved, this will trigger transcription of a tox gene in the green population. The tox gene encodes for a toxin, which will kill cells of the green population to some extent (they should not all be killed). As soon as the density of the green population decreases, also concentration of B will drop off. Since B is vitally necessary for the red population, a drop of B will lead to a decline in cell density of the red population. This will affect the concentration of A in the medium, thus enabling the green population to re-grow. Along with growth, the pool of B will be replenished, thus allowing density of the red population to increase. When the threshold concentration of A will be reached, toxin expression in the green pop will be triggered, etc etc etc etc etc....
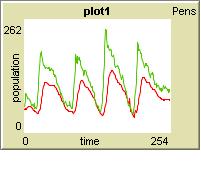
Simulation
I used an agent-based simulation program called netlogo. For those who don't know what an agent-based simulation is: The special thing is that you have different agents (each e.coli would be an agent in our case) that act quasi-concurrently. The simulation program runs one time step after another. You can now tell all the agents seperately what they shall do during each time step (e.g. to produce some A, or to die when theres too much toxG around).
At the beginning of the simulation, I set both popolations to 50, A to 50 and B to 100. Both populations start to grow and produce A and B respectively, until A reaches its threshold value of 140 and the green start to die. The red continue to grow for some time until B is consumed, then they start to die as well. When A falls below the threshold, the green start to grow again, producing more B, which in turn leads to the growth of the red.
Problems: Both populations have their peaks at approximately the same time and are, therefore, in phase. I don't know whether this behavior is realistic, because I don't know enough about biology to simulate the rising and falling concentration of A and B realistically.
I've collected the various questions whose answers would help me to make the simulation much more realistic:
- What's the concentration of A and B in the beginning?
- How many reds and greens are there initially?
- How much A/B is produced by every red/green bacteria during each timestep?
- How do the red bacteria react (i.e. how fast do they procreate/die) to rising/falling concentrations (and possibly distributions) of B?
- What's the threshold of A? Does it depend only on the concentration or also on the geographical distribution?
- How fast do the green bacteria die (through toxG) when A surpasses its threshold?
- How fast do the red and green ones procreate (in relation to emulating A or B, respectively)?
- How fast do A and B 'wear off' with time?
Discussion
>> for comments, questions and temporary remarks go to the Talk:Predator-Prey_Behavior
Back to Development Group.
Back to ETH Zurich main page.