Tokyo Alliance: Significance
From 2006.igem.org
(Difference between revisions)
(→Our contributions to the iGEM and SynBio World) |
|||
| Line 15: | Line 15: | ||
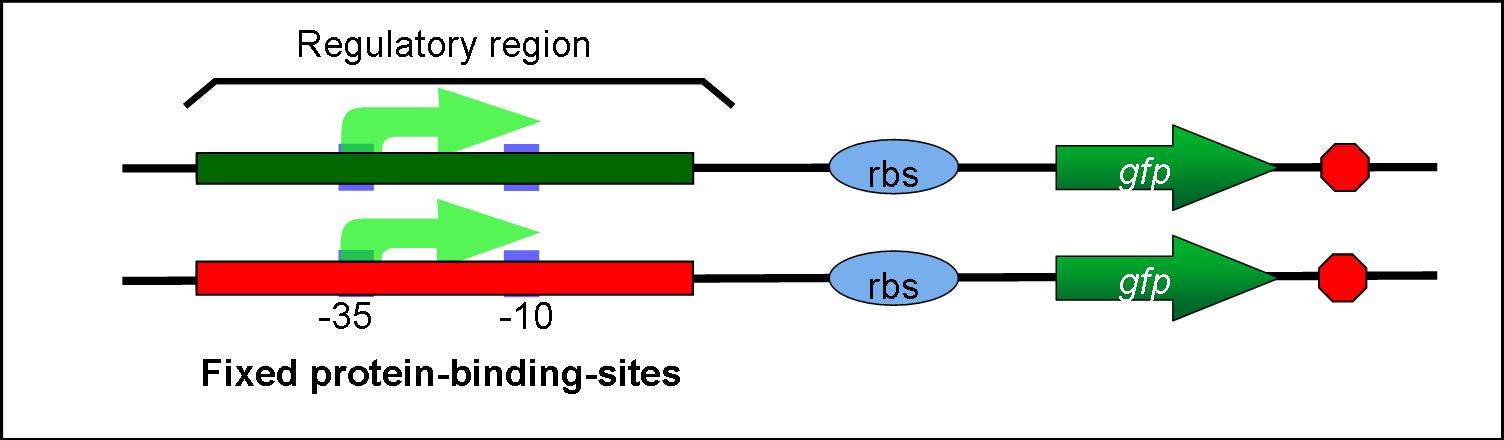
:As long as we use previous iGEM methods, it is arduous to construct regulatory regions to which multiple transcriptional regulators bind. | :As long as we use previous iGEM methods, it is arduous to construct regulatory regions to which multiple transcriptional regulators bind. | ||
| - | |||
:[[Image:significance2.jpg|500px|]] | :[[Image:significance2.jpg|500px|]] | ||
| Line 21: | Line 20: | ||
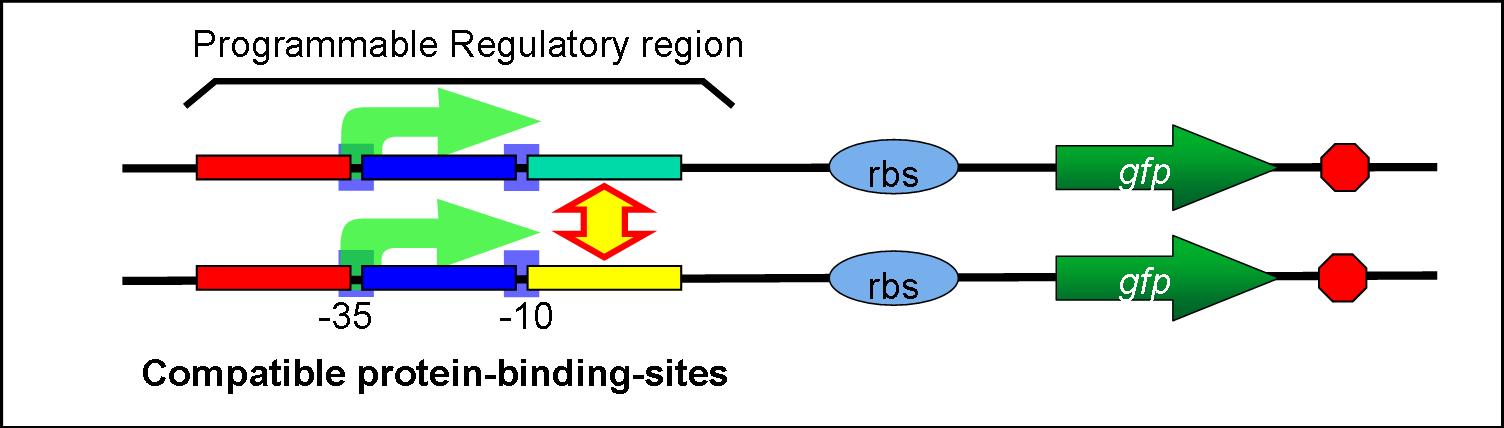
:Combinatorial rewiring of these parts allows replacing regulators. | :Combinatorial rewiring of these parts allows replacing regulators. | ||
| - | :For example, if so, we can replace the regulator binding sites | + | :For example, if so, we can replace the regulator binding sites between -10 and RBS. |
| + | |||
'''See our [[Tokyo_Alliance:_Conclusion#Construction_Family_Tree |Construction Family Tree]] (Results Page) !''' | '''See our [[Tokyo_Alliance:_Conclusion#Construction_Family_Tree |Construction Family Tree]] (Results Page) !''' | ||
Latest revision as of 13:04, 1 November 2006
Our contributions to the iGEM and SynBio World
- Expansion of inducer library.
- One of the biggest problem in Synthetic Biology is only a few numbers of inducer of transcriptional regulators.
- Diverse regulatory region library by Systematic design/construction
- As long as we use previous iGEM methods, it is arduous to construct regulatory regions to which multiple transcriptional regulators bind.
- Combinatorial rewiring of these parts allows replacing regulators.
- For example, if so, we can replace the regulator binding sites between -10 and RBS.
See our Construction Family Tree (Results Page) !